Mesenchymal stem cell-macrophages influence cancer stemness and AKT1/YKL-39 expression in triple-negative breast cancer
- Department of Biomedical Science, Advanced Medical and Dental Institute, Universiti Sains Malaysia, Bertam, 13200 Kepala Batas, Pulau Pinang, Malaysia
- Preparatory Centre for Science and Technology, Universiti Malaysia Sabah, Jalan UMS, 88400 Kota Kinabalu, Sabah, Malaysia
- Breast Cancer Translational Research Program (BCTRP), Advanced Medical and Dental Institute, Universiti Sains Malaysia, Bertam, 13200 Kepala Batas, Pulau Pinang, Malaysia
- Department of Toxicology, Advanced Medical and Dental Institute, Universiti Sains Malaysia, Bertam,13200 Kepala Batas, Pulau Pinang, Malaysia
- Integrative Pharmacogenomics Institute (iPROMISE), Universiti Teknologi MARA Selangor, Puncak Alam Campus, 42300 Puncak Alam, Selangor, Malaysia
Abstract
Introduction: Triple-negative breast cancer (TNBC) has greater infiltration of tumor-associated macrophages (TAMs) than other breast-cancer subtypes. Mesenchymal stromal/stem cells (MSCs) have been shown to shape macrophage polarization, promoting the conversion of naïve M0 macrophages to an M1-like phenotype that restricts cancer-cell growth. However, reciprocal relationships between macrophages and cancer stem cells (CSCs) may promote the CSC niche and drive macrophages to transition toward an M2-like phenotype in the tumor microenvironment (TME). This study explores the effects of MSC-polarized macrophages (M0/MSCs) on CSCs and evaluates their role in TNBC progression.
Methods: M0 macrophages derived from THP-1 cells were co-cultured with UC-MSCs for 72 h, then co-cultured with breast cancer stem cells (BCSCs) originating from MDA-MB-231 cells. Gene-expression profiles were assessed by qRT-PCR of total RNA extracted from each cell type, focusing on M1/M2 polarization and CSC markers. Subsequently, MDA-MB-231 and MCF-10A cells were cultured for 72 h in conditioned medium (CM) obtained from the CSC/M0-MSC co-culture. An MTT assay was used to measure cell viability, and the impact on breast-cancer-related genes was analyzed by qRT-PCR.
Results: Co-culturing M0/MSCs with BCSCs (CSC/M0-MSCs) induced an M2-like macrophage phenotype, marked by elevated IRF4 and increased TNF-α expression. Conversely, SOX-2 and OCT-4 expression was reduced in BCSCs co-cultured with M0/MSCs. MDA-MB-231 cells exposed to co-culture-derived CM for 72 h showed a non-significant decrease in viability and down-regulation of AKT1 and YKL-39.
Conclusion: Co-culturing M0/MSC macrophages with BCSCs elevated IRF4 and TNF-α in macrophages while reducing SOX-2 and OCT-4 in BCSCs. The elevation of TNF-α indicates M2-like polarization; however, its context-dependent effects on stemness warrant further investigation. In addition, CM from the CSC/M0-MSC co-culture modulated AKT1 and YKL-39 expression. These findings underscore the need to further elucidate MSC–macrophage–BCSC interactions to develop more effective therapies targeting the TNBC TME.
Introduction
Breast cancer (BC) is the primary cause of death and the most widespread malignancy in females worldwide1. The GLOBOCAN 2020 report shows that BC overtook lung cancer as the most frequently occurring cancer globally, with 2.3 million new documented cases, accounting for 11.7 % of all cancer cases. Despite its high incidence, BC ranks fifth in female cancer mortality2. Based on the expression of vital indicators such as the human epidermal growth factor receptor 2 (HER2), progesterone receptor (PR), and estrogen receptor (ER), the disease is typically classified into several subtypes1. Among these subtypes, triple-negative breast cancer (TNBC) constitutes roughly 15–20 % of total BC cases3. TNBC is associated with the poorest prognosis due to its aggressive nature, characterized by high-grade tumors with rapid proliferation. Moreover, the treatment options are limited due to the lack of HER2 overexpression and minimal ER and PR expression (<1%)3. Consequently, TNBC is primarily treated through chemotherapy, radiotherapy, and surgical interventions4. However, these therapeutic approaches often have limited success, highlighting the need for more effective treatments.
The tumor microenvironment (TME) exerts a strong influence on TNBC growth and immune response. The complex cellular and soluble network within the TME, encompassing adipocytes, endothelial cells, fibroblasts, immune cells, and their secretions, contributes to tumor progression5. Notably, compared with conventional breast cancer subtypes, TNBC tumors exhibit higher secretion of vascular endothelial growth factor (VEGF) and a higher percentage of tumor-associated macrophages (TAMs), which drive tumor cell invasion and proliferation6. TAMs, which are formed from circulating monocytes, play a major role in shaping the TME. These macrophages are capable of transforming into alternatively activated (M2) or classically activated (M1) macrophages, determined by the signals they receive7. M1 macrophages are typically activated by colony-stimulating factor (CSF), tumor necrosis factor (TNF), and interferon (IFN), and are associated with tumor suppression. They express surface markers such as CD80, CD86, and CD40, along with pro-inflammatory cytokines like IL-6, TNF-α, and IL-1β8. Conversely, M2 macrophages are linked to tissue repair and immune suppression and are triggered by cytokines such as TGF-β, IL-4, and IL-10. They express markers such as CD206, CD163, and CD209 and release anti-inflammatory molecules8. TAMs are often classified as M2-like due to their immune-suppressive characteristics, though they exhibit plasticity and can display features of both M1 and M2 phenotypes in response to the TME9, 10. This plasticity has made repolarizing TAMs towards an M1-like phenotype a promising therapeutic approach for treating cancer.
Mesenchymal stromal/stem cells (MSCs) are capable of regulating immune responses through various mechanisms, including their interaction with macrophages. MSCs can influence macrophage polarization, often promoting the M2-like phenotype, which augments wound healing and tissue regeneration11. However, in the context of cancer, an M2 phenotype may not be advantageous. Recent research has indicated that MSCs can promote the M1-like phenotype in naïve macrophages (M0) and later polarize them to an M2-like state upon further stimulation12, 13, 14, 15. Our previous work corroborated these findings, demonstrating that MSCs can enhance M1-like polarization in macrophages, as indicated by increased levels of IRF5 and reduced expression of M2 genes when co-cultured with M0 macrophages. Research targeting the MSC–macrophage axis in TNBC remains limited; however, our study also demonstrated that conditioned medium (CM) from the M0/MSC co-culture significantly hindered the growth of MDA-MB-231 cells by downregulating AKT1 and YKL-39 genes16, 17.
A group of tumor cells with stem-like characteristics, known as cancer stem cells (CSCs), is key to the establishment, growth, and spread of tumors. CSCs can alter the composition of the TME by secreting extracellular vesicles (EVs) and various factors such as growth factors, metabolites, chemokines, and cytokines18. These factors contribute to the establishment of a unique niche that supports CSC self-renewal and proliferation. In the TME, the association between CSCs and macrophages is especially crucial for maintaining the CSC niche. CSCs attract macrophages to the TME, where they are converted into TAMs, which, in turn, create a supportive microenvironment for CSC survival and stemness19. The secretion of TGF-β, IL-13, and IL-4 by CSCs promotes the M2-like phenotype in macrophages20. Given that MSCs can induce M1 polarization in macrophages, an important question is whether MSC-polarized macrophages can maintain an M1-like phenotype even in the presence of CSCs and whether this might reduce M2 polarization.
This study seeks to determine the effect of MSC-polarized macrophages (M0/MSC) on breast cancer stem cells (BCSCs), particularly with regard to macrophage polarization and their impact on breast cancer cell behavior. The findings may provide insight into the interactions among MSCs, macrophages, and BCSCs, paving the way for the design of innovative therapies targeting the TME of TNBC.
Methods
Culture of THP-1, MDA-MB-231, UC-MSCs, and MCF-10A cells
THP-1 cells were cultivated in RPMI-1640 medium containing L-glutamine and supplemented with 1 % Pen-Strep and 10 % FBS. MDA-MB-231 cells and UC-MSCs were grown in DMEM/F12 medium supplemented with sodium bicarbonate, L-glutamine, 1 % Pen-Strep, and 10 % FBS. DMEM/F12 medium comprising L-glutamine, sodium bicarbonate, 20 ng mL⁻¹ epidermal growth factor (EGF), 1 % Pen-Strep, and 10 % FBS was used to culture MCF-10A cells. All cells were maintained at 37 °C in a humidified 5 % CO incubator.
THP-1 cells were incubated with 20 nM PMA for 48 h to differentiate them into naïve (M0) macrophages. M0 macrophages were co-cultured with UC-MSCs in 6-well plates (Corning, USA) for 30 h, separated by 0.4 µm-pore transwell inserts (Corning, USA). The insert contained 3.5 × 10⁴ UC-MSCs, whereas the well contained 3.5 × 10⁵ M0 macrophages. This ratio was selected from a previous study by Vasandan .16. Four co-culture conditions were established for 30 h: M0 macrophages (M0), LPS-stimulated M0 macrophages (LPS-M0), M0 macrophages co-cultured with UC-MSCs (M0-MSCs), and LPS-stimulated M0 macrophages co-cultured with UC-MSCs (LPS-M0-MSCs). To maintain cell viability, all co-cultures were performed in complete medium containing the same FBS concentration as the corresponding monocultures, consistent with previous investigations21.
Enrichment of mammospheres from MDA-MB-231 cells
MDA-MB-231 cells were used to generate mammospheres following the protocol of Wang .22. Cells (1 × 10⁵) were seeded in ultra-low-attachment (ULA) plates in mammosphere medium consisting of DMEM/F12 supplemented with sodium bicarbonate, L-glutamine, 0.4 % FBS23 and 1 % Pen-Strep. Additionally, the medium was supplemented with 20 ng mL⁻¹ basic fibroblast growth factor (bFGF), insulin, 20 ng mL⁻¹ EGF, and 2 % B-27. The ULA plates were incubated at 37 °C in 5 % CO. An initial volume of 2 mL medium per well was used, and 0.5 mL fresh medium was added every 3 days. Non-adherent spherical clusters (mammospheres) appeared after 5 days and continued to grow for up to 14 days. Mammospheres > 50 µm were counted microscopically on days 5, 7, 10, and 14, and mammosphere-forming efficiency (MFE) was calculated as the percentage of spheres relative to seeded cells. Mammospheres were dissociated with 0.25 % trypsin-EDTA for 5 min at room temperature, and the resulting single cells were analysed for CSC-associated markers (CD44 and CD24) by flow cytometry. RNA was also extracted to quantify SOX-2 and OCT-4 expression. After 14 days of enrichment, the cells were co-cultured with M0/MSCs to examine their effect on macrophage polarization.
Flow-cytometry analysis of enriched mammospheres
After PBS washing, mammospheres were enzymatically dissociated to obtain a single-cell suspension. Cells (1 × 10⁵) were resuspended in 100 µL BD Pharmingen™ Stain Buffer containing monoclonal antibodies against human CD44-PE and CD24-APC (R&D Systems, USA) and incubated for 30 min at 4 °C in the dark. Unlabelled cells served as negative controls. Cells were then washed with 1 × PBS to remove unbound antibodies. Stained samples were acquired on a BD FACSCalibur and analysed with BD FACSComp software to determine CD44 and CD24 expression.
Co-culture of enriched BCSCs with M0/MSCs
Enriched BCSCs (1 × 10⁵ cells well⁻¹) were seeded in 6-well ULA plates. M0-MSC macrophages (1 × 10⁵ cells well⁻¹) were added to 0.4 µm transwell inserts (Corning, USA) and co-cultured with BCSCs at a 1:1 ratio for 72 h, allowing exchange of soluble factors while preventing direct contact. All co-cultures were performed in complete medium containing FBS, as in previous studies21. To preserve their individual requirements, BCSCs (lower chamber) were maintained in 0.4 % FBS mammosphere medium, whereas M0-MSC macrophages (upper chamber) were kept in 10 % FBS medium. After 72 h, total RNA was isolated from both cell populations. Macrophages were analysed for M1 and M2 polarization markers, and BCSCs for the pluripotency genes OCT-4 and SOX-2. The conditioned medium (CM) collected from the co-culture was used in subsequent experiments with MDA-MB-231 and MCF-10A cells.
Treatment of MDA-MB-231 and MCF-10A cells with CM from the BCSC/M0-MSC co-culture
To assess the effect of CM on cell proliferation, MDA-MB-231 or MCF-10A cells (5 × 10³ cells well⁻¹) were seeded in 96-well plates for 24 h; the medium was replaced with 100 µL CM from the BCSC/M0-MSC co-culture, and the cells were incubated for 72 h prior to the MTT assay.
For gene-expression studies, MDA-MB-231 or MCF-10A cells (3.5 × 10⁵ cells well⁻¹) were seeded in 6-well plates for 24 h. The medium was then replaced with a 1:1 mixture of CM and fresh medium, and cells were incubated for 72 h. Total RNA was extracted and cDNA synthesised for analysis of AKT1, mTOR, p53, and YKL-39 expression.
MTT assay
After 72 h stimulation with CM, the medium was removed and 10 µL MTT solution plus 90 µL fresh medium were added to each well. Cells were incubated for 3 h at 37 °C, 5 % CO, after which 100 µL DMSO was added to dissolve the formazan crystals. Absorbance was read at 570 nm with an ELISA microplate reader after a 10 min incubation.
Total RNA isolation and cDNA synthesis
Cellular RNA was extracted with TRIsure™ reagent (Bioline, UK) following the manufacturer’s protocol. RNA quality and quantity were assessed with a NanoDrop spectrophotometer (Thermo Fisher Scientific, USA). cDNA was generated with the Tetro cDNA Synthesis Kit (Bioline, UK) according to the manufacturer’s instructions.
Quantitative real-time PCR
Relative mRNA levels were quantified on a Real-Time PCR System (Applied Biosystems, USA) using the SensiFAST SYBR Hi-ROX Kit (Bioline, UK) in triplicate. Each 20 µL reaction contained 6.4 µL nuclease-free water, 0.8 µL 10 µM primer mix, 10 µL 2 × SensiFAST SYBR Hi-ROX Master Mix and 2 µL cDNA.
Sequences of the primers used for q-PCR analyses
|
Gene |
Forward (5’-3’) |
Reverse (5’-3’) |
|---|---|---|
|
|
AATCCCATCACCATCTTCCA |
TGGACTCCACGACGTACTCA |
|
|
TTCTCTCCTGGGCTGTCTCTG |
CTATACAGCTAGGCCCCAGGG |
|
|
GCTGATCGACCAGATCGACAG |
CGGTTGTAGTCCTGCTTGC |
|
|
CCTCTCTCTAATCAGCCCTCTG |
GAGGACCTGGGAGTAGATGAG |
|
|
TACGGCGCTCGTCATCGATTT |
TAGAGTCGCCACCCTGATGT |
|
|
GAAGGACGGGAGCAGGCGGC |
CCTCCTCCAGGCAGCCCT |
|
|
AGTGGACCAGTGGAAACAGG |
TTCAGCGATGTCTTGTGAGG |
|
|
AAGATGACCTTGCTGCCT |
TGATCTAAGAGGAAGTCAGG |
|
|
CCCCTCCATCCTTTCTTCTC |
ATGAGCCAGATCAGGGACTG |
|
|
GTTGATCCTCGGACCTGGCTA |
GGTTGCCTCTCACTCGGTTCT |
|
|
GCCGAGTGGAAACTTTTGTCG |
GCAGCGTGTACTTATCCTTCTT |
Statistical analysis
Unless otherwise stated, all data are presented as the mean ± standard deviation (SD) of at least three independent experiments. Analyses were performed with SPSS. One-way ANOVA was used for comparisons involving more than two groups; Tukey’s post-hoc test was applied when variances were equal and Games–Howell when variances were unequal (Levene’s test). Student’s independent t-test was used for two-group comparisons. Statistical significance was set at P < 0.05 (*), P < 0.01 (**), and P < 0.001 (***).
Results
Mammosphere formation enriches for BCSCs derived from MDA-MB-231 cells
After 14 days under CSC-specific culture conditions, MDA-MB-231 cells generated more mammospheres > 50 µm in diameter, exhibiting loose, grape-like structures (Figure 1A–B). Although not statistically significant, the mammosphere-forming efficiency (MFE) increased from 0.0067 % ± 0.0012 % on day 5 to 0.0143 % ± 0.0042 % on day 14, indicating enhanced stem-like properties over time (Figure 1B).
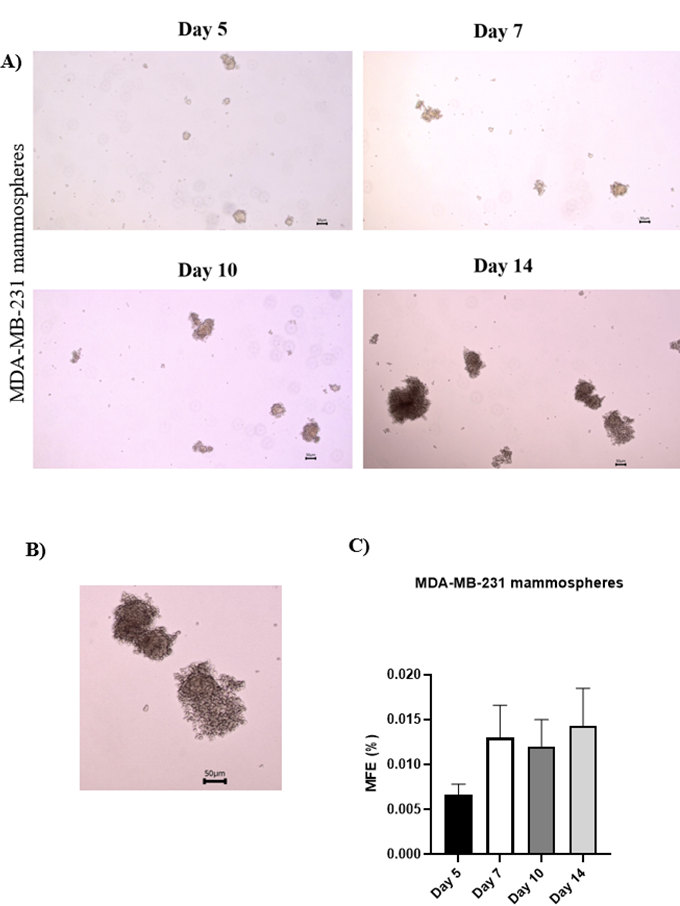
Formation of mammospheres from enriched BCSCs derived from MDA-MB-231 cells for 14 days. A) Morphology of mammospheres from MDA-MB-231 cells at day 5, day 7, day 10 and day 14 of culture taken at 40X magnification. Scale bar: 50 µm. B) Enlarged image showing a cropped region of a representative day 14 mammosphere from the image in (A). Scale bar: 50 µm. C) Bar graph displaying the differences in mammosphere forming efficiencies (MFEs) enriched from MDA-MB-231 cells at day 5, day 7, day 10 and day 14. Data are shown as mean ± SD of three independent experiments (n = 3). Statistical significance was determined using one-way ANOVA followed by Tukey’s post hoc test for multiple comparisons. Error bars denote standard deviation (SD).
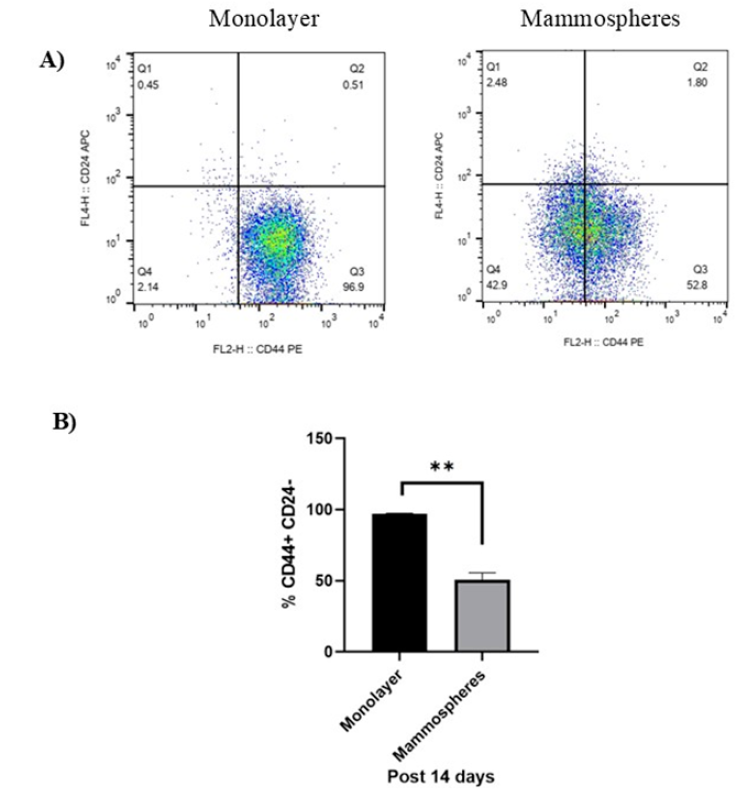
Mammospheres are enriched for CD44⁺CD24⁻/low cells
At day 14, the proportion of CD44CD24 cells was assessed. Monolayer cultures contained 97.3 % ± 0.29 % CD44CD24 cells, whereas mammospheres contained 50.7 % ± 4.21 % (Figure 2A). Thus, mammosphere culture significantly reduced this population (P < 0.01) but it remained > 50 % (Figure 2B).
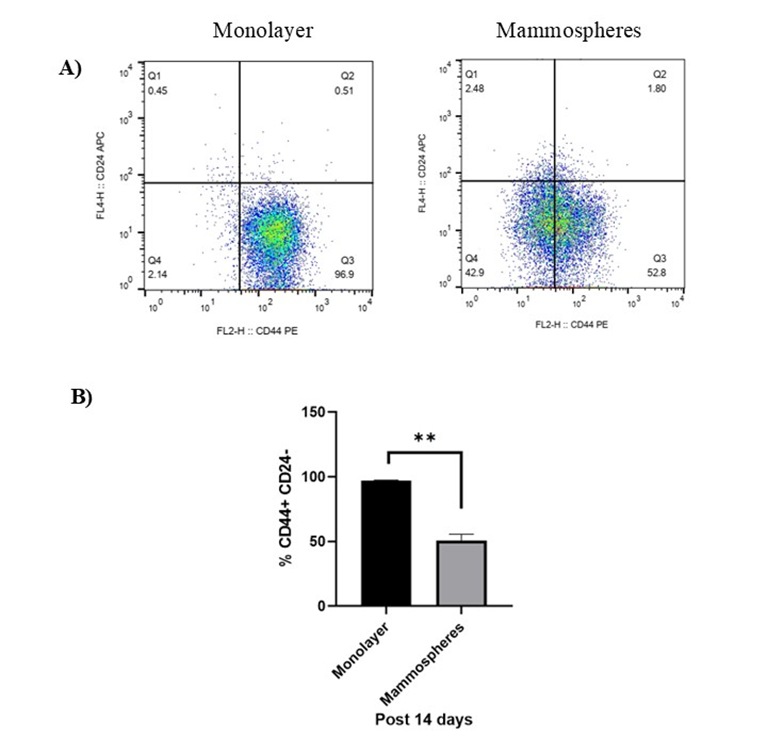
Flow cytometry analysis of CD44+CD24-/low surface markers expression in MDA-MB-231 cells monolayer and enriched mammospheres for 14 days. A) Dot plot and B) bar graph of CD44+CD24-/low surface markers expression in monolayer and enriched mammospheres of MDA-MB-231 cells. Data are shown as mean ± SD of three independent experiments (n=3). Statistical analysis was determined using student's independent t-test. Error bars denote standard deviation (SD). Asterisks indicate significance: **P < 0.01.
SOX-2 and OCT-4 expression is elevated in mammospheres derived from MDA-MB-231 cells
SOX-2 and OCT-4 mRNA levels were quantified in MDA-MB-231 monolayers and in first- and second-generation mammospheres. First-generation spheres were collected after 14 days, whereas second-generation spheres were produced by re-seeding 1 × 10⁵ cells well⁻¹ and culturing for 72 h. Although changes did not reach statistical significance, both SOX-2 (Figure 3A) and OCT-4 (Figure 3B) were higher in first- and second-generation spheres than in monolayers.
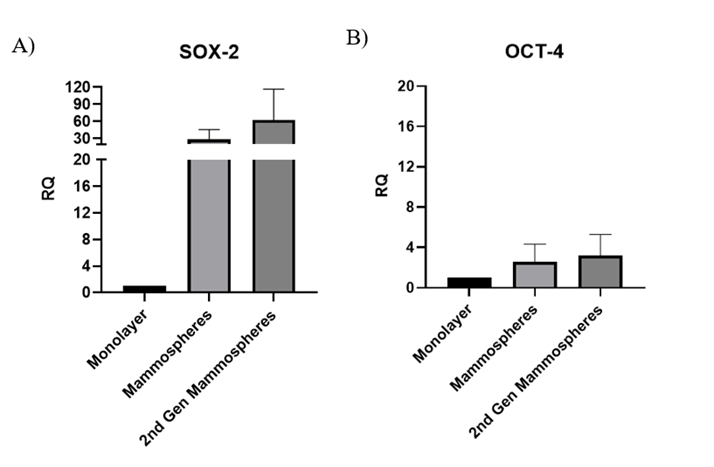
Pluripotency genes expression in monolayer of MDA-MB-231 cells and their derived mammospheres post 14 days. Bar chart of A) SOX-2 and B) OCT-4 relative fold genes expression in monolayer of MDA-MB-231 cells and their derived mammospheres. All relative expressions were quantified and normalised to GAPDH. Data are shown as mean ± SD of three independent experiments (n=3). Statistical significance was determined using one-way ANOVA followed by Tukey’s multiple comparison test. Error bars denote standard deviation (SD).
M0-MSCs and LPS-primed M0-MSCs up-regulate IRF4, whereas only M0-MSCs increase TNF-α after co-culture with BCSCs
Expression of M1-associated (IRF5, TNF-α) and M2-associated (IRF4, IL-10) genes was analysed in untreated M0 macrophages and in M0- and LPS-M0-MSCs after 72 h of co-culture with BCSCs. IRF5 and IL-10 were unchanged (Figure 4A, D). Co-culture with M0-MSCs significantly increased TNF-α versus co-cultures containing M0 or LPS-M0 cells (P < 0.05; Figure 4B). Both M0-MSCs and LPS-M0-MSCs displayed higher IRF4 than untreated M0 cells (P < 0.01 and P < 0.001, respectively; Figure 4C).
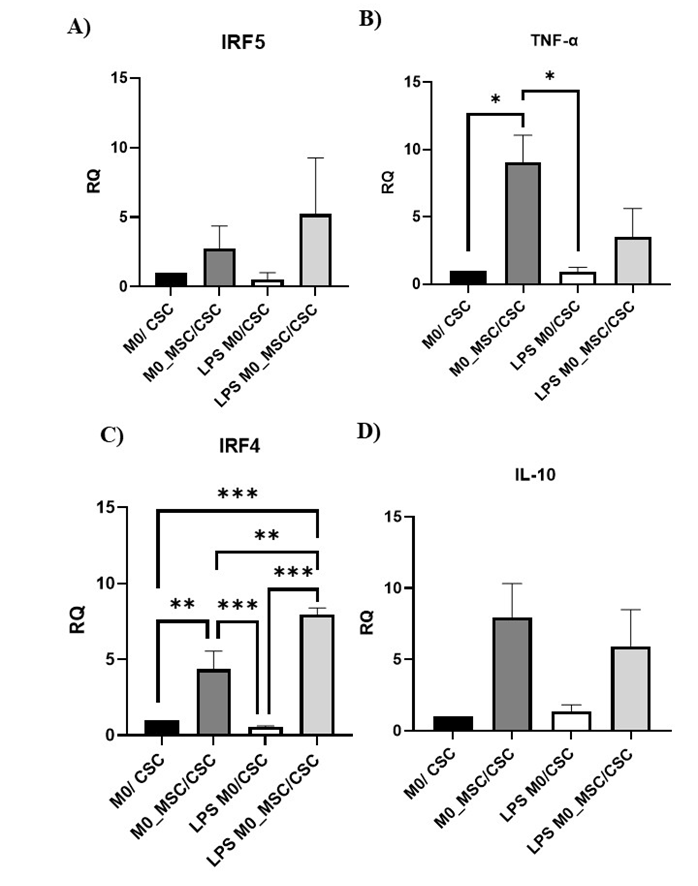
Expression of M1/M2-associated genes following co-culture of M0-MSC and LPS M0-MSC macrophages (1×105 cells) with BCSCs (1×105 cells) for 72 hours. M0-MSC and LPS M0-MSC macrophages were co-cultured for 72 hours and total RNA was extracted and reverse transcribed into cDNA. Relative quantification of M1-associated genes A) IRF5, B) TNF-α and M2-associated genes, C) IRF4, D) IL-10 expression was determined by qRT-PCR and normalized to GAPDH. Data are shown as mean ± SD of three independent experiments (n = 3). Statistical significance was determined using one-way ANOVA followed by Tukey’s test for equal variances or Games-Howell test when variances were unequal. Error bars denote standard deviation (SD). Asterisks indicate significance: *P < 0.05, **P < 0.01, ***P < 0.001.
M0-MSCs reduce SOX-2 and OCT-4 expression in BCSCs
Co-culture of BCSCs with either M0-MSCs or LPS-M0 cells significantly decreased SOX-2 relative to BCSCs alone (both P < 0.05; Figure 5A). OCT-4 was likewise reduced in BCSCs cultured with M0-MSCs (P < 0.01) or LPS-M0 cells (P < 0.05) (Figure 5B).
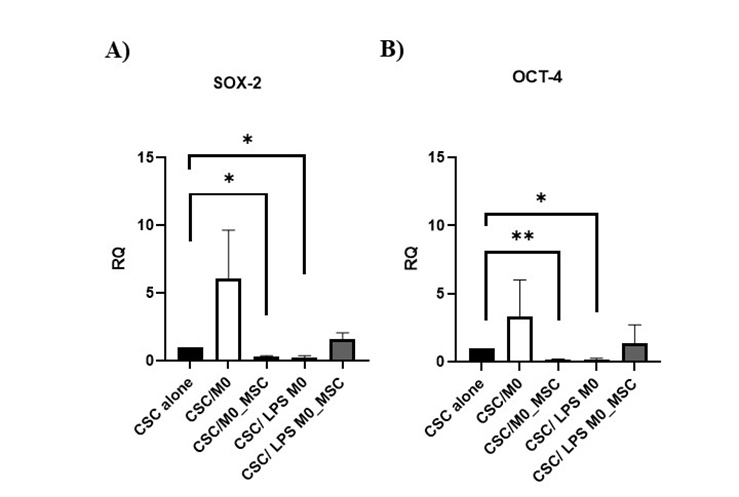
Expression of pluripotency genes in BCSCs (1×10⁵ cells) following co-culture with M0-MSC and LPS M0-MSC macrophages (1×10⁵ cells) for 72 hours. The total RNA of BCSCs were extracted and reverse transcribed into cDNA. Relative quantification of A)
Conditioned medium (CM) from M0-MSC/BCSC co-cultures down-regulates p53, YKL-39 and AKT1 in MDA-MB-231 cells
After 72 h in CM from M0-MSC/BCSC co-cultures (CSC/M0-MSCs), MDA-MB-231 viability fell to 91.83 % ± 3.08 %, but this reduction was not significant (Figure 6A). AKT1 was significantly lower in cells treated with CM from CSC/M0, CSC/M0-MSCs or CSC/LPS-M0-MSCs than in fresh medium controls (all P < 0.01; Figure 6B). YKL-39 expression was likewise reduced by CM from CSC/M0 (P < 0.05), CSC/M0-MSCs (P < 0.01) and CSC/LPS-M0-MSCs (P < 0.05; Figure 6C). mTOR was unaffected (Figure 6D). Finally, p53 was down-regulated by CM from BCSCs alone and from CSC/M0-MSCs (both P < 0.05; Figure 6E).
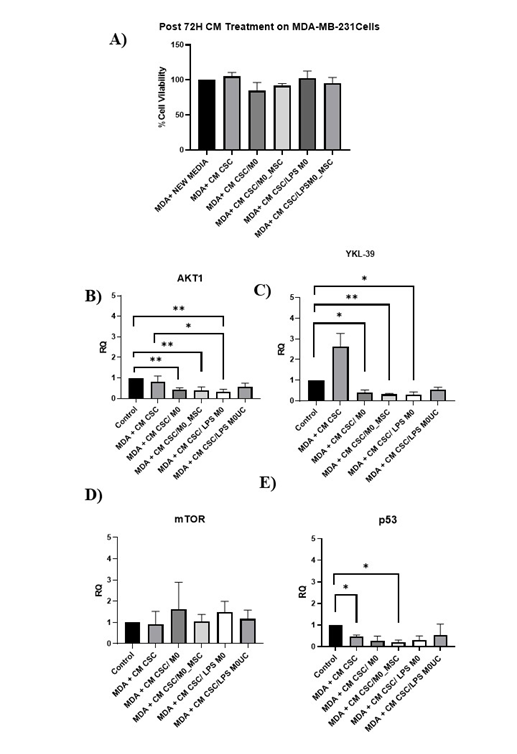
The proliferation and expression of
Conditioned medium from M0-MSC/BCSC co-cultures does not alter proliferation or or expression in MCF-10A cells
Proliferation of MCF-10A cells was unchanged by CM from M0-MSC/BCSC co-cultures (Figure 7A). In contrast, CM from CSC/LPS-M0-MSCs significantly reduced proliferation relative to untreated cells and to CM from BCSCs alone (both P < 0.01). CM from CSC/M0-MSCs did not affect or in MCF-10A cells (Figure 7B–E).
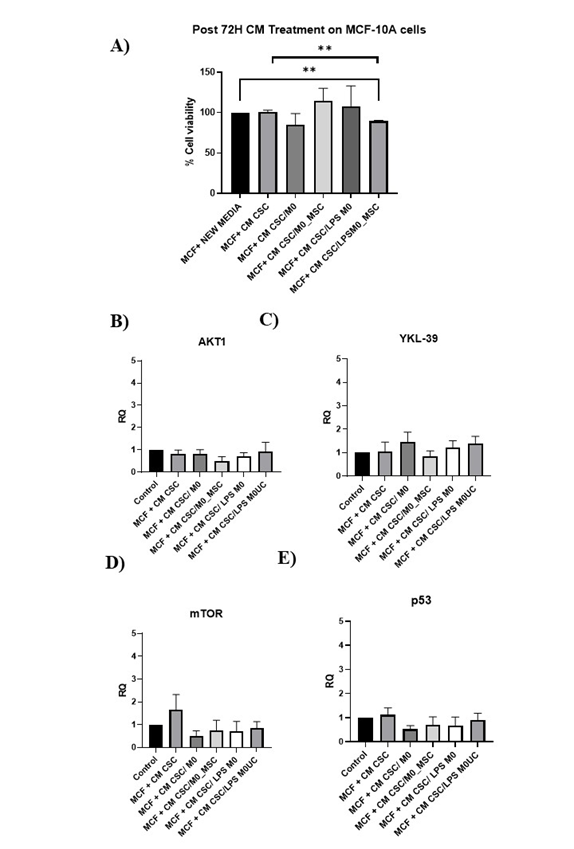
The proliferation and expression of AKT1, YKL-39 and p53 genes in MCF-10A cultured in CM (1:1 ratio with fresh media) from untreated and treated M0 co-cultured with BCSCs for 72 hours. A) Proliferation rate of MCF-10A cultured in CM from aforementioned conditions, the expression of B)
Discussion
MDA-MB-231 cells are a commonly utilized TNBC cell line, with 19,331 citations in PubMed24. They are extensively employed as a model for TNBC because they lack HER2, PR, and ER, making them an appropriate representation in several studies25, 26, 27. However, reliance on a single cell line may limit the extent to which the results can be applied. According to Conner ., the tumorigenic and metastatic potential of distinct TNBC cell lines varies; MDA-MB-468, MDA-MB-231, and BT-549 cells, for instance, are thought to be extremely tumorigenic and metastatic. Therefore, it is recognized that utilizing a broader panel of TNBC cell lines with different metastatic potential would reflect TNBC variability and increase the robustness of experimental findings24.
The formation of mammospheres is a strategy employed to enrich cultures with BCSCs. BCSCs are grown in a non-adherent, serum-deprived environment in the presence of EGF and bFGF to promote CSC proliferation. When breast-cancer cells are grown in non-adherent conditions, they form multicellular three-dimensional (3D) spheres known as mammospheres. Wang . found that the MFE (diameter > 50 µm) from enriched BCSCs of primary breast tumors and MCF-7 cells was higher than that of MDA-MB-231 cells, possibly owing to a link between estrogen-receptor expression and mammosphere formation22. Our mammosphere-MFE data showed no significant differences over time, although there was a clear upward trend that peaked on Day 14. Jardin discovered that MDA-MB-231 cells could produce mammospheres with the help of Orai1α and Orai1β28. Additionally, the percentage of CD44CD24 cells was lower in mammospheres than in monolayer cells, although SOX-2 and OCT-4 levels were elevated. The fraction of CD44CD24 cells, however, was still high and remained above 50%. A reduced fraction of CD44CD24 cells may be associated with phenotypic plasticity and marker variability in CSCs. Primary breast tumors and MCF-7 cells were previously found to exhibit a markedly elevated percentage of CD44/CD24ESALin cells under mammosphere culture conditions, whereas MDA-MB-231 cells already had a high number of CD44/CD24ESALin (96.2 % ± 3.6 %) before enrichment22. A later study by Huang . reported that MDA-MB-231 mammospheres exhibited a reduced percentage of CD44/CD24 cells (53.48 % ± 4.99 %) compared with parental monolayer cells (97.25 % ± 1.74 %), a trend consistent with our findings29. This implies that the expression of these markers may be modulated by mammosphere culture conditions. Yousefnia proposed that CD44⁺ is not a universal BCSC marker because of diversity between breast-cancer subtypes. However, stemness-associated genes such as OCT-4 and Nanog were significantly up-regulated in mammospheres from MCF-7, SKBR3, and MDA-MB-231, demonstrating that stemness was maintained or strengthened despite surface-marker variability30. OCT-4, NANOG, and SOX-2 are stem-cell markers and are recognised to have crucial roles in regulating embryonic-stem-cell pluripotency and self-renewal, thereby driving tumor development and differentiation31.
The influence of co-culturing BCSCs with MSC-modulated macrophages was then assessed. According to most research, TAMs can aid CSCs and their niche, and CSCs can stimulate macrophage activation and polarization in a way that enhances tumor growth32. However, less is currently known about reciprocal interactions between CSCs and macrophages33. In our study, MSC-modulated macrophages (M0-MSC) co-cultured with BCSCs for 72 h were found to have increased expression of IRF4, a gene associated with M2 macrophages, compared with M0 macrophages in the presence of BCSCs. Deng . previously discovered that co-culture of the murine macrophage cell line RAW264.7 with ovarian CSCs (OCSCs) reduced mRNA levels of M1 markers, while M2 markers such as IL-10 were increased relative to untreated RAW264.7 cells. Deng . conducted the initial study to show that OCSCs activate the PPARγ/NF-κB pathway, leading to macrophage M2 polarization34. PPARγ suppresses NF-κB, blocking the activation of pro-inflammatory genes, including IL-2, IL-6, IL-8, metalloproteinases, and TNF-α34.
Interestingly, in our study, M0-MSC macrophages showed higher TNF-α expression than M0 and LPS-primed M0 macrophages, despite evidence that BCSCs enhance a pro-tumoral phenotype in macrophages. M1 macrophages generate high amounts of TNF-α, which induces cell death in the TME, whereas M2 macrophages produce TNF-α that promotes epithelial-to-mesenchymal transition (EMT) and stemness in cancer35. Notably, Kratochvill . revealed that TNF is required for reducing the number of pro-tumoral M2 macrophages36. The sequential exposure of macrophages to MSCs and subsequently BCSCs demonstrates the dynamic nature of macrophage polarization. Previously, we found that MSCs inhibit TNF-α but favour a shift to an M1-like phenotype (elevated IRF5 and reduced M2-associated genes) in naïve macrophages17. Subsequent interaction with BCSCs could restore TNF-α expression while also increasing the M2-associated marker IRF4, reflecting a hybrid phenotype. Interestingly, Chen found that co-culture of M2-TAMs with SMMC-7721 hepatocellular-carcinoma cells led to increased TNF-α37.
The TME promotes reprogramming by providing an environment rich in lactic acid, lipids, and cytokines, as well as hypoxia, low pH, and glucose deprivation38. By encouraging both immunosuppressive and inflammatory responses simultaneously, this hybrid phenotype may promote tumor development. Additionally, our work showed that when LPS-primed M0-MSC macrophages were co-cultured with BCSCs (LPS M0-MSC/CSC), their IRF4 expression was higher than that of LPS M0 macrophages, enhancing their M2-like phenotype. Raghavan. reported that direct interaction between M0 macrophages and OCSCs increased the M2 marker CD206 compared with unsorted ovarian-cancer cells33. The elevation of CD206 was further supported by enhanced IL-10 expression in CSCs, and the study suggests that paracrine WNT activation contributes to these pro-tumoral characteristics33.
Lipopolysaccharide (LPS) was used to promote M1-like polarization in macrophages to generate a pro-inflammatory baseline. This system enables the study of BCSCs’ ability to alter macrophage polarization, possibly shifting them toward an M2-like phenotype with enhanced IRF4 production. Such a change could help explain how BCSCs contribute to an immunosuppressive TME. For example, Piao . discovered that exosomes derived from MDA-MB-231 TNBC cells caused macrophage polarization toward the M2 phenotype, as evidenced by elevated CD206 expression both and 39. The outcomes of our research imply that tumor-derived factors can override early polarization states to promote an immunosuppressive environment conducive to tumor growth, underlining the relevance of investigating the effect of BCSCs on LPS-primed macrophages.
Expression of the cancer-stem-cell markers OCT-4 and SOX-2 in BCSCs was assessed after 72 h of contact with M0-MSC–derived macrophages. BCSCs co-cultured with M0-MSC–derived macrophages showed lower SOX-2 and OCT-4 expression than the control (BCSCs cultured alone). Cytokines secreted by TAMs, such as FGF, TGF-β1 and PGE2, can help differentiated cancer cells regain colony-forming CSC traits and maintain stemness in tumors40. A breast-cancer model showed that TAMs stimulate the EGFR/STAT3/SOX2 pathway and enhance expression of ABCG2, OCT-4, SCA-1, NANOG and SOX-2, all typical CSC markers41.
TNF-α regulates CSCs by promoting BCSC expansion through the NF-κB/HIF-1α/Slug pathway35. Although TNF-α can drive tumor development and metastasis, it may also exert opposing, tumor-inhibitory effects on CSCs. Our findings showed that interaction with M0-MSC–derived macrophages induced an M2-like macrophage phenotype but did not increase CSC-like traits in BCSCs.
Despite its well-described anti-tumor properties, TNF-α enhances T47D-cell growth while exerting pro-apoptotic, anti-mitogenic effects in MCF-7 cells42, 43. TNF-α can also influence CSCs by activating EMT pathways, increasing sphere formation, up-regulating stem-cell markers and promoting tumor progression44. Conversely, Abdolvand reported that a 1-week TNF-α treatment reduced CXCR4 expression and the BCSC population in MDA-MB-231 cells45. These contrasting findings may explain the down-regulation of stemness markers observed in our study.
Conditioned medium (CM) from BCSC/M0-MSC co-cultures (CSC/M0-MSCs) was collected and applied to MDA-MB-231 or MCF-10A cells to evaluate its effects. After 72 h of treatment, MDA-MB-231 proliferation decreased slightly; however, the change was not significant. CM from CSC/M0-MSCs significantly down-regulated AKT1 and YKL-39 in MDA-MB-231 cells compared with both the control (cells cultured alone) and cells treated with CM from untreated BCSCs. Up to 70 % of breast cancers display PI3K/AKT dysregulation, and AKT up-regulation correlates with poor prognosis46. AKT1 suppresses apoptosis and promotes cell-cycle progression via p27, p21 and cyclin D146.
YKL-39 (chitinase-3-like protein 2, CHI3L2) exhibits monocyte-chemotactic and pro-angiogenic activities and is expressed by M2 macrophages in renal, glioma and breast cancers47. It is also found in the tumor mesenchyme, cytoplasm and nucleus of gastric-cancer cells, where it is linked to angiogenesis and disease progression46. Its role in TNBC remains unclear. Both YKL-39 and its homolog YKL-40 are chitinase-like proteins (CLPs)48; knock-down of YKL-40 reduced proliferation, invasion and migration of HEC-1A endometrial-cancer cells49. Similar YKL-39-silencing studies could clarify its contribution to TNBC.
Our previous work showed that CM from M0/MSC co-cultures strongly inhibited MDA-MB-231 cells by down-regulating AKT1 and YKL-3917. Although CM from CSC/M0-MSCs did not markedly affect cell growth, the observed suppression of AKT1 and YKL-39 suggests indirect modulation of oncogenic signalling via inflammatory mediators.
Co-culture of MSC-modulated macrophages (M0-MSCs) with breast cancer stem cells (BCSCs) increases TNF-α expression and produces conditioned medium (CM) that suppresses AKT1 and YKL-39 expression in MDA-MB-231 cells. These findings suggest that TNF-α influences AKT1 and YKL-39 expression in tumor cells. However, the direct involvement of TNF-α in regulating these genes has not been confirmed and warrants further investigation, particularly for YKL-39, whose significance in TNBC remains poorly defined48. Despite the dual function of TNF-α reported in the literature, most studies link elevated TNF-α with enhanced tumor growth, invasion, and stemness, particularly in TNBC. TNF-α activates MMP9 expression via PI3K/Akt and p42/p44 MAPK pathways and also increases ZEB2 expression, an EMT regulator in TNBC cell lines50. Notably, Wu found that TNF-α improved chemotherapy sensitivity and radiation efficacy in the breast cancer cell lines MCF-7, MDA-MB-231, and ZR-75-1 by altering the NF-κB pathway, demonstrating that TNF-α can have anti-tumor effects in specific contexts51. TNF-α’s influence on tumor development depends on activation of its receptors, TNFR1 and TNFR252. TNF-α primarily induces apoptosis through TNFR1, which is expressed in all cell types, and can also promote cell survival depending on TNF-α levels and co-present cytokines in the microenvironment35. In contrast, TNFR2 is mainly expressed on immune cells and has been linked to accelerated tumor progression35. Both receptors signal through NF-κB pathways, but their effects differ according to receptor expression levels and competition with signalling molecules such as TRAF253. The literature indicates that TNFR2 may modulate tumor immunoregulation and represents a potential therapeutic target53. Zhao demonstrated that TNFR2 promotes colorectal cancer progression through the phosphoinositide-3-kinase/AKT pathway54. Pretreatment with a TNFR2-neutralizing antibody effectively inhibited the PI3K/AKT and ERK signaling cascades in cholangiocarcinoma cells55. Complex I, which comprises TNFR2, TNF-associated factors 1, 2, and 3, and cellular apoptosis inhibitors 1 and 2, activates NF-κB, MAPK, and AKT signaling pathways to promote cell survival42. Although TNFR2 may influence AKT1, this remains speculative because there is no direct evidence. Consequently, the potential contribution of TNFR2 suppression to the down-regulation of AKT1 in MDA-MB-231 cells cultured with CM from CSC/M0-MSC should be investigated further. Future studies should also explore TNF-α cytokine profiling and pathway inhibition to clarify its role in CSC–macrophage-mediated tumor modulation.
In contrast, CM from CSC/M0-MSCs had no inhibitory effect on the normal breast cell line MCF-10A, whereas CM from CSC/LPS-stimulated M0-MSCs did. However, CM from CSC/M0-MSCs did not significantly alter AKT1, YKL-39, mTOR, or p53 expression in MCF-10A cells. According to Lee , indirect co-culture of MCF-10A cells with TAMs promotes their transition to a pre-cancerous phenotype. CCL2, or monocyte chemoattractant protein-1, possesses potent chemotactic activity to recruit monocytes and macrophages and can promote tumor metastasis, invasion, and immune evasion56, 57. Elevated MMP expression and activity, particularly MMP-9, have been linked to multiple cancers and poorer patient outcomes58. After co-culture with TAMs, MCF-10A cells exhibit increased activation of JNK1/2, ERK1/2, p38 MAPK, and Akt pathways55. The findings of Lee . differ from ours, as we observed an inhibitory effect, rather than enhanced growth, when MCF-10A cells were treated with CM from CSC/LPS-M0-MSCs. Moreover, this CM did not significantly change AKT1 expression in MCF-10A cells.
While this study provides useful insights, several limitations should be acknowledged. First, we enriched BCSCs using low-adherence culture supplemented with EGF and bFGF, a well-established approach for mammosphere formation. Enrichment was confirmed by assessing the stem-cell markers CD44CD24 and the pluripotency factors SOX-2 and OCT-4, which are typically elevated in CSCs59, 60. However, functional assays such as ALDH activity, side-population analysis, or colony-formation assays were not performed and would strengthen BCSC validation in future studies60. Second, THP-1-derived macrophages may not fully recapitulate the complexity of primary human macrophages, particularly for M2 polarization60. Shiratori reported that only PBMC-derived macrophages displayed strong M2a induction in response to IL-461. Conversely, primary monocyte-derived macrophages present practical limitations, including limited proliferative capacity, the need for freshly isolated cells in every experiment, an inability to be cryopreserved, and substantial donor-to-donor variability, all of which can affect experimental outcomes62. Third, batch- and donor-to-donor variability in the commercial UC-MSCs used may alter their immunomodulatory capacity, including macrophage polarization, thereby affecting reproducibility. Additionally, although qRT-PCR provides valuable data on M1/M2 marker expression, protein-level analyses (., ELISA or flow cytometry) were not performed and would provide more direct evidence of functional polarization. Finally, our study sought to model the TME by allowing MSC-modulated macrophages to interact with cancer cells. However, future experiments using CM from MSC-only and macrophage-only cultures will help delineate the individual contributions of each cell type and clarify how MSC-educated macrophages influence BCSC behaviour. Despite these limitations, our study advances understanding of the MSC–macrophage–BCSC triad and may inform the development of targeted therapies aimed at the TME in TNBC.
Conclusion
Our results indicate that when BCSCs are grown as mammospheres in serum-free conditions, they exhibit enhanced stem cell characteristics, as shown by increase trend in MFE and elevated levels of OCT-4 and SOX-2 expression, despite a decrease in the CD44CD24 population. Co-culturing of BCSCs with M0/MSC macrophages induced an M2-like macrophage phenotype, marked by elevated IRF4 and TNF-α levels, while the stemness markers in BCSCs were reduced. Additionally, CM from the CSC/M0-MSCs co-culture downregulated AKT1 and YKL-39 genes expression. The findings indicate that, while CSC/M0-MSCs secreted factors did not significantly hinder MDA-MB-231 cell growth, they may modulate anti-tumor pathways by downregulating AKT1 and YKL-39 expression.
Abbreviations
BC: Breast Cancer, BCSC: Breast Cancer Stem Cell, CSC: Cancer Stem Cell, ER: Estrogen Receptor, HER2: Human Epidermal Growth Factor Receptor 2, MSC: Mesenchymal Stromal/Stem Cell, PR: Progesterone Receptor, TAM: Tumor-Associated Macrophage, TNBC: Triple-Negative Breast Cancer, UC-MSC: Umbilical Cord Mesenchymal Stromal/Stem Cell, M0: Naïve macrophage, M1: Pro-inflammatory macrophage, M2: Anti-inflammatory macrophage, THP-1: Human monocytic cell line, MDA-MB-231: Triple-negative breast cancer cell line, MCF-10A: Non-tumorigenic breast epithelial cell line, AKT1: AKT Serine/Threonine Kinase 1, bFGF: Basic Fibroblast Growth Factor, CD24: Cluster of Differentiation 24, CD44: Cluster of Differentiation 44, CHI3L2: Chitinase-3-Like Protein 2, CSF: Colony-Stimulating Factor, EGF: Epidermal Growth Factor, EMT: Epithelial-to-Mesenchymal Transition, EV: Extracellular Vesicle, FGF: Fibroblast Growth Factor, GAPDH: Glyceraldehyde-3-Phosphate Dehydrogenase, IFN: Interferon, IL-1β: Interleukin 1 Beta, IL-4: Interleukin 4, IL-6: Interleukin 6, IL-10: Interleukin 10, IL-13: Interleukin 13, IRF4: Interferon Regulatory Factor 4, IRF5: Interferon Regulatory Factor 5, LPS: Lipopolysaccharide, mTOR: Mammalian Target of Rapamycin, OCT-4: Octamer-Binding Transcription Factor 4, PGE2: Prostaglandin E2, SOX-2: SRY-Box Transcription Factor 2, TGF-β: Transforming Growth Factor Beta, TNF: Tumor Necrosis Factor, TNF-α: Tumor Necrosis Factor Alpha, VEGF: Vascular Endothelial Growth Factor, YKL-39: Chitinase-3-Like Protein 2, YKL-40: Chitinase-3-Like Protein 1, ANOVA: Analysis of Variance, BCA: Bicinchoninic Acid Assay, cDNA: Complementary DNA, CM: Conditioned Medium, DMSO: Dimethyl Sulfoxide, ELISA: Enzyme-Linked Immunosorbent Assay, FACS: Fluorescence-Activated Cell Sorting, FBS: Fetal Bovine Serum, MFE: Mammosphere-Forming Efficiency, MTT: 3-(4,5-Dimethylthiazol-2-yl)-2,5-Diphenyltetrazolium Bromide, PBS: Phosphate-Buffered Saline, PMA: Phorbol 12-Myristate 13-Acetate, qRT-PCR: Quantitative Real-Time Polymerase Chain Reaction, SD: Standard Deviation, ULA: Ultra-Low Attachment, 3D: Three-Dimensional, Fig: Figure, h: Hour(s), min: Minute(s), mL: Milliliter, µL: Microliter, nm: Nanometer, µm: Micrometer, ng: Nanogram, nM: Nanomolar, P: p-value, Pen-Strep: Penicillin-Streptomycin, TME: Tumor Microenvironment
Acknowledgments
None.
Author’s contributions
The research study was conceptualized by RM, MAY, BHY, MYA, and MSR. Experimental work was carried out by NRJ. Data analysis was carried out by NRJ, RM, BHY and MSR. The manuscript was drafted by RM, NRJ, MAY and MYA. RM, as the principal investigator, led the study funded by the Research University Individual and Special Research grant. All authors carefully evaluated and endorsed the final manuscript for submission to the journal.
Funding
This research was financed by the Research University Individual Grant (1001/CIPPT/8012328) and the Research University Grant for BCTRP (1001/CIPPT/8070033), both provided by Universiti Sains Malaysia.
Availability of data and materials
Data and materials associated with this study are available upon request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Declaration of generative AI and AI-assisted technologies in the writing process
The authors utilized OpenAI’s ChatGPT to support the refinement of language, organization of content, and formatting throughout the development of this manuscript. All outputs generated were carefully reviewed and modified by the authors to ensure accuracy and integrity. The authors accept full responsibility for the final content presented.
Competing interests
The authors declare that they have no competing interests.

