Functional Divergence of Human Lung BDCA1⁺ and BDCA3⁺ Dendritic Cells at Rest and in Response to Viral Stimuli
- Molecular Biology and Genetics Program, Department of Basic Sciences and Humanities, Faculty of Arts and Sciences, Cyprus International University (CIU), Via Mersin 10, Nicosia, Northern Cyprus, Türkiye
Abstract
Dendritic cells (DCs) play a pivotal role in shaping immune responses in the human lung; however, the subset-specific transcriptional programs that underpin their steady-state and stimulus-driven functions remain incompletely defined. In this validation-oriented, integrative in silico study, we analyzed microarray data from dataset GSE43184 to profile immune-gene expression in the two principal human lung DC subsets, BDCA1⁺ (cDC2) and BDCA3⁺ (cDC1) cells. Rather than serving as a standalone discovery resource, GSE43184 was used illustratively to validate and synthesize existing knowledge of DC subset divergence. Analysis of baseline expression revealed that BDCA1⁺ DCs exhibit markedly higher levels of pro-inflammatory cytokines and chemokines—including IL1B, CXCL8, CCL5, and TNF—whereas BDCA3⁺ DCs displayed lower overall expression but relatively higher levels of interferon-stimulated genes (ISGs) such as ISG15 and IFIT1. Principal-component analysis and hierarchical clustering further confirmed the distinct transcriptional identities of the two subsets. Targeted functional enrichment of genes up-regulated in BDCA1⁺ cells indicated predominant activation of inflammatory and cytokine-signalling pathways. Comparative validation with public single-cell RNA-seq datasets and prior studies employing viral stimuli (e.g., RSV, poly I:C) revealed concordant subset-specific responses: BDCA1⁺ DCs are inflammation-prone, whereas BDCA3⁺ DCs initiate ISG-dominated antiviral programmes. A conceptual summary and curated literature synthesis reinforced these functional distinctions. Collectively, this study provides a synthesis-driven validation analysis that illustrates the functional divergence between BDCA1⁺ and BDCA3⁺ dendritic cells, rather than a de novo gene-discovery effort. Our findings support the view that BDCA1⁺ and BDCA3⁺ DCs are intrinsically programmed for distinct immunological roles within the lung microenvironment and respond differentially to viral cues.
Introduction
Dendritic cells (DCs) constitute a critical nexus between innate and adaptive immunity, functioning not only as antigen-presenting cells but also as orchestrators of immune responses against a wide range of pathogens across diverse tissues 1,2. In the lung, a frontline organ continuously exposed to airborne agents, DCs play an especially crucial role in maintaining immune surveillance and balancing defense with tolerance 3. Human conventional DCs (cDCs) are broadly classified into two major subsets: BDCA1 (CD1c, also known as cDC2), which are proficient T-helper-cell priming, and BDCA3 (CD141, or cDC1), recognized for their cross-presentation capacity and contribution to antiviral immunity 2,4,5.
Recent evidence indicates that these DC subsets are transcriptionally and functionally specialized, with implications for infection, tissue homeostasis, and immune-mediated disease 3,5. Systematic transcriptional profiling by microarray and single-cell RNA sequencing (scRNA-seq) has revealed distinct gene-expression patterns that differentiate BDCA1 and BDCA3 subsets within the lung microenvironment. Single-cell atlases such as the Human Lung Cell Atlas (HLCA) and PanglaoDB consistently show that BDCA1 DCs express higher levels of pro-inflammatory cytokines and chemokines (IL1B, CXCL8, CCL5), whereas BDCA3 DCs display elevated interferon-stimulated genes (ISG15, IFIT1), suggesting complementary and non-redundant functional programs that enhance immune readiness 3,6.
Both in vitro and in vivo studies support this dichotomy. During RSV infection, BDCA1 DCs secrete high concentrations of pro-inflammatory cytokines (IL1B, IL6, TNFα), whereas BDCA3 DCs exhibit attenuated inflammatory responses but increased expression of co-inhibitory markers (PD-L1, IL-10) 7,8. Conversely, BDCA3 DCs excel in type I interferon signalling and antigen cross-presentation following TLR3 stimulation (e.g., poly I:C) 5,7, underscoring their differential predisposition towards antiviral defence mechanisms.
Despite these insights, comprehensive comparative transcriptomic analyses that integrate steady-state and stimulus-specific profiles of BDCA1 and BDCA3 lung DCs remain scarce. Specifically, studies systematically analysing baseline immune-gene expression, integrating pathway-level assessments, and validating cell-type specificity with single-cell data and functional inference are lacking. A more granular understanding of these transcriptional landscapes is essential for devising targeted immunotherapies and for interpreting DC dynamics in respiratory disease.
In this validation-oriented, synthesis-driven study, we analysed a publicly available microarray dataset (GSE43184) of human lung-derived BDCA1 and BDCA3 DCs to consolidate and refine existing transcriptomic observations. Our aims were threefold: (1) to characterise baseline immune-gene expression profiles across selected cytokines, chemokines, and interferon-stimulated genes; (2) to delineate global transcriptional architecture through principal-component analysis (PCA), hierarchical clustering, and Reactome-based functional enrichment; and (3) to validate these observations by comparative analysis with single-cell RNA-seq atlases and prior studies employing canonical viral stimuli (RSV and poly I:C).
Rather than proposing new mechanistic discoveries, this work integrates multiple data sources to validate and contextualise known differences between BDCA1 and BDCA3 DCs. Key findings include elevated resting expression of inflammatory cytokines in BDCA1 DCs, distinct clustering patterns separating the two subsets, and enrichment of inflammatory and interferon pathways consistent with the existing literature. By emphasising synthesis and validation, our analysis underscores the reproducible transcriptomic polarisation of human lung DC subsets and highlights their relevance for understanding DC-driven immunity in infection, vaccination, and immunotherapeutic settings.
Methods
Data Acquisition
The microarray dataset analyzed in this study was retrieved from the Gene Expression Omnibus (GEO) (accession no. GSE43184). The dataset comprises transcriptomic profiles of two human lung dendritic-cell subsets—BDCA1 and BDCA3 cells—purified from healthy donors. Both raw CEL files and the processed series matrix, together with the corresponding annotation file for the Affymetrix Human Genome U133 Plus 2.0 Array (GPL570), were downloaded to enable accurate probe-to-gene mapping.
Pre-processing and Normalization
Data pre-processing was performed in the R statistical environment using Bioconductor packages. Raw CEL files were processed with the Robust Multi-array Average (RMA) algorithm, which performs background correction, quantile normalization and log transformation. Probes were mapped to gene symbols using the GPL570 annotation file; probes lacking valid gene assignments or mapping ambiguously to multiple genes were excluded. When several probes corresponded to the same gene, the probe with the highest mean expression across all samples was retained to represent that gene in downstream analyses.
Differential Expression Analysis
Samples were stratified into BDCA1 and BDCA3 dendritic-cell subsets according to the dataset metadata. Differential expression analysis was performed with the limma package, which fits linear models to each gene and applies empirical Bayes moderation to stabilize variance estimates. Significance was defined by a false-discovery-rate (FDR)-adjusted p-value < 0.05 (Benjamini–Hochberg method) and an absolute log fold-change ≥ 1. Genes meeting these criteria were deemed differentially expressed and prioritized for subsequent interpretation. Given the limited sample size (n = 2 per subset), all findings should be regarded as hypothesis-generating. Complete differential-expression statistics for key cytokine and interferon-stimulated genes are provided in Supplementary Table S1.
Functional Enrichment Analysis
The biological significance of transcriptional differences was examined with the clusterProfiler package. Gene Ontology enrichment was assessed across biological-process, molecular-function and cellular-component categories, whereas Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was used to identify over-represented signaling cascades. Enrichment terms with an adjusted p-value < 0.05 were considered significant. To provide immunological context, curated immune gene sets were retrieved from the ImmPort and InnateDB databases. Expression of cytokines, chemokines and interferon-stimulated genes was evaluated to highlight subset-specific immune functions.
Unsupervised Analyses
Global transcriptional patterns were interrogated using unsupervised approaches. Principal-component analysis (PCA) was conducted with the prcomp function in R, and results were visualized to illustrate the clustering of samples by dendritic-cell subset. Hierarchical clustering was performed on the 1,000 most variable genes using Euclidean distance and complete linkage. Heat-maps generated with the pheatmap package confirmed the distinct transcriptional signatures of BDCA1 and BDCA3 cells.
Cross-validation with Public Datasets
To evaluate the robustness of our findings, results were cross-validated with external datasets. Single-cell RNA-sequencing data from the Human Lung Cell Atlas were queried to corroborate the transcriptional signatures of BDCA1 and BDCA3 subsets. Additionally, published studies examining dendritic-cell responses to respiratory syncytial virus (RSV) infection and polyinosinic:polycytidylic acid (poly I:C) stimulation served as independent validation resources. This comparative analysis confirmed the inflammation-prone transcriptional identity of BDCA1 cells and the interferon-stimulated-gene dominance in BDCA3 cells.
Data Visualization and Reproducibility
All figures were generated in R using the ggplot2, EnhancedVolcano and pheatmap packages. Volcano plots summarized differential expression (Supplementary Figure S1); PCA plots illustrated sample clustering; and heat-maps depicted expression profiles of immune-related genes. Analyses were executed in a Linux-based computational environment. All scripts are available from the corresponding author upon reasonable request, and GSE43184 is publicly accessible through the GEO repository.
Results
Baseline Immune Gene Expression in Human Lung DC Subsets
Immune gene expression profiles of human lung-resident BDCA1 and BDCA3 dendritic cells were analysed using microarray data retrieved from the Gene Expression Omnibus (GEO; accession GSE43184). The analysis centred on a curated panel of cytokines, chemokines and interferon-stimulated genes (ISGs) typically linked to innate immune activation and antiviral defences. This gene subset was chosen to elucidate the baseline immunological predisposition of each dendritic-cell subset.
BDCA1 dendritic cells demonstrated significantly higher basal expression of the pro-inflammatory mediators IL1B, CXCL8, CCL5 and TNF than did BDCA3 cells. As these cytokines orchestrate leukocyte recruitment, potentiate inflammation and initiate early antiviral responses, the data indicate that BDCA1 cells are transcriptionally primed for vigorous inflammatory activity. Conversely, BDCA3 cells exhibited lower and more homogeneous expression of the same genes, consistent with a more restrained inflammatory phenotype at rest (Figure 1; Supplementary Figure S1; Supplementary Table S1). Collectively, these findings highlight intrinsic subset-specific contributions to pulmonary immune surveillance and early host defence.
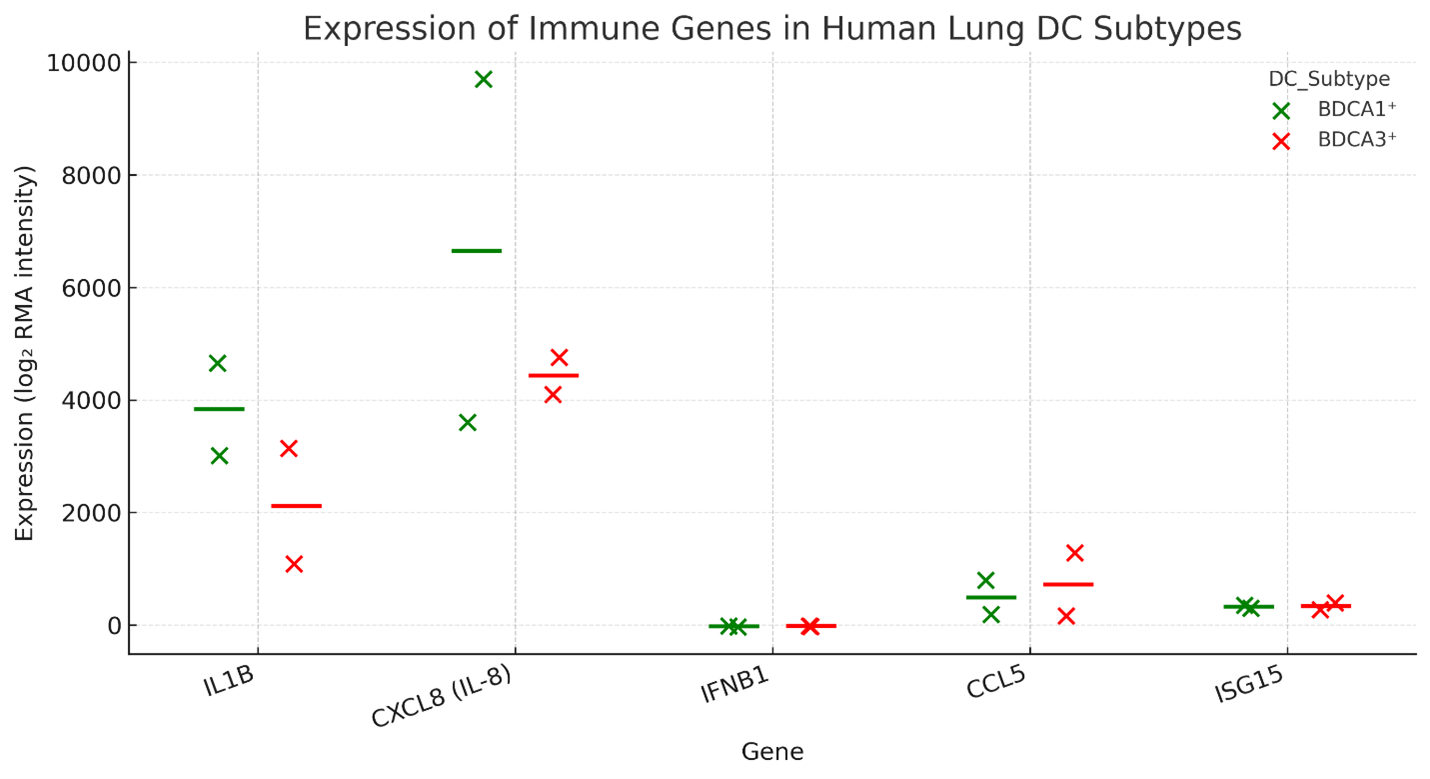
Baseline immune gene expression in human lung dendritic cell subsets. Dot-plot representation of RMA-normalized log2 expression intensities for selected immune-related genes (
Clustering and Dimensional Reduction Reveal Subset-Specific Expression Signatures
To delineate global transcriptional differences, we applied principal component analysis (PCA) to the immune-gene expression matrix. This dimensionality-reduction method separated BDCA1 and BDCA3 samples along principal component 1 (PC1), which explained the largest proportion of variance (Figure 2A). The distinct clusters indicate that each subset harbours a discrete immune-gene signature even under steady-state conditions. Nevertheless, the limited sample size (n = 2 per subset) warrants caution, as technical variability may partially account for the observed segregation.
Unsupervised hierarchical clustering provided complementary confirmation of this segregation. BDCA1 cells clustered tightly and displayed concerted up-regulation of hallmark inflammatory genes (IL1B, CXCL8 and CCL5), whereas BDCA3 cells formed a discrete cluster with uniformly lower transcript levels (Figure 2B). These observations corroborate that BDCA1 and BDCA3 lung dendritic cells constitute transcriptionally distinct populations endowed with differential immune-activation potential.
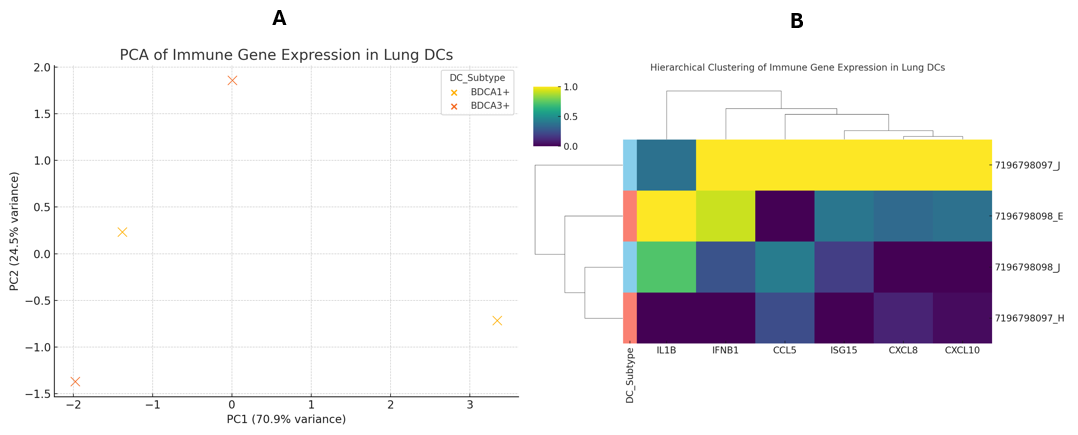
Transcriptional separation of lung dendritic cell subsets by immune gene expression.(A) Principal component analysis (PCA) of normalized expression values for 15 immune-related genes (cytokines, chemokines, ISGs) in BDCA1+ (CD1c+) and BDCA3+ (CD141+) human lung dendritic cells. Each point represents one biological replicate (n = 2 per subset). The first principal component (PC1) accounts for the majority of variance and clearly segregates the two DC subsets. (B) Unsupervised hierarchical clustering heatmap of the same gene set across individual samples. Expression values were row-standardized (z-score) to highlight relative differences. Columns represent samples (annotated by subset: BDCA1+ vs. BDCA3+), rows represent genes, and dendrograms reflect clustering by Euclidean distance and complete linkage. Abbreviations: PCA, principal component analysis; PC1, principal component 1; ISGs, interferon-stimulated genes.
Functional Enrichment Shows Cytokine-Driven Signatures in BDCA1 Dendritic Cells
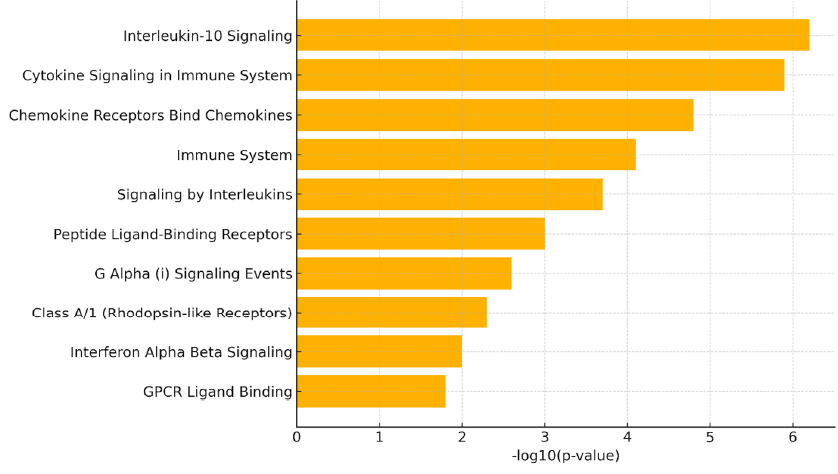
To interrogate the functional consequences of these transcriptional disparities, we conducted pathway-enrichment analysis with the Reactome database. The analysis was confined to the six genes most strongly up-regulated in BDCA1 cells relative to BDCA3 cells—CCL5, CXCL8, ISG15, IFNB1, CXCL10 and IL1B—which were consistently elevated across biological replicates and are integral to innate immunity. Notably, this approach constitutes a focused interrogation of biological processes linked to the top cytokine- and interferon-related genes, rather than a genome-wide, unbiased screen.
The Reactome analysis demonstrated significant enrichment of several immune pathways (Figure 3). The most prominent terms included Interleukin-10 signalling, Cytokine signalling in the immune system and Chemokine receptors bind chemokines, each pivotal for leukocyte trafficking, immunoregulation and antiviral defence. Additional enriched categories—Interferon-α/β signalling, G-protein-coupled receptor (GPCR) ligand binding and Signalling by interleukins—underscore the multifaceted immunological capacity of BDCA1 dendritic cells at rest. Collectively, these data suggest that BDCA1 lung dendritic cells are transcriptionally poised to mount broad-spectrum immune responses, especially during inflammation or viral infection. Nevertheless, the enrichment analysis is inherently descriptive and limited to pathways already annotated for the curated cytokine- and interferon-related gene set.
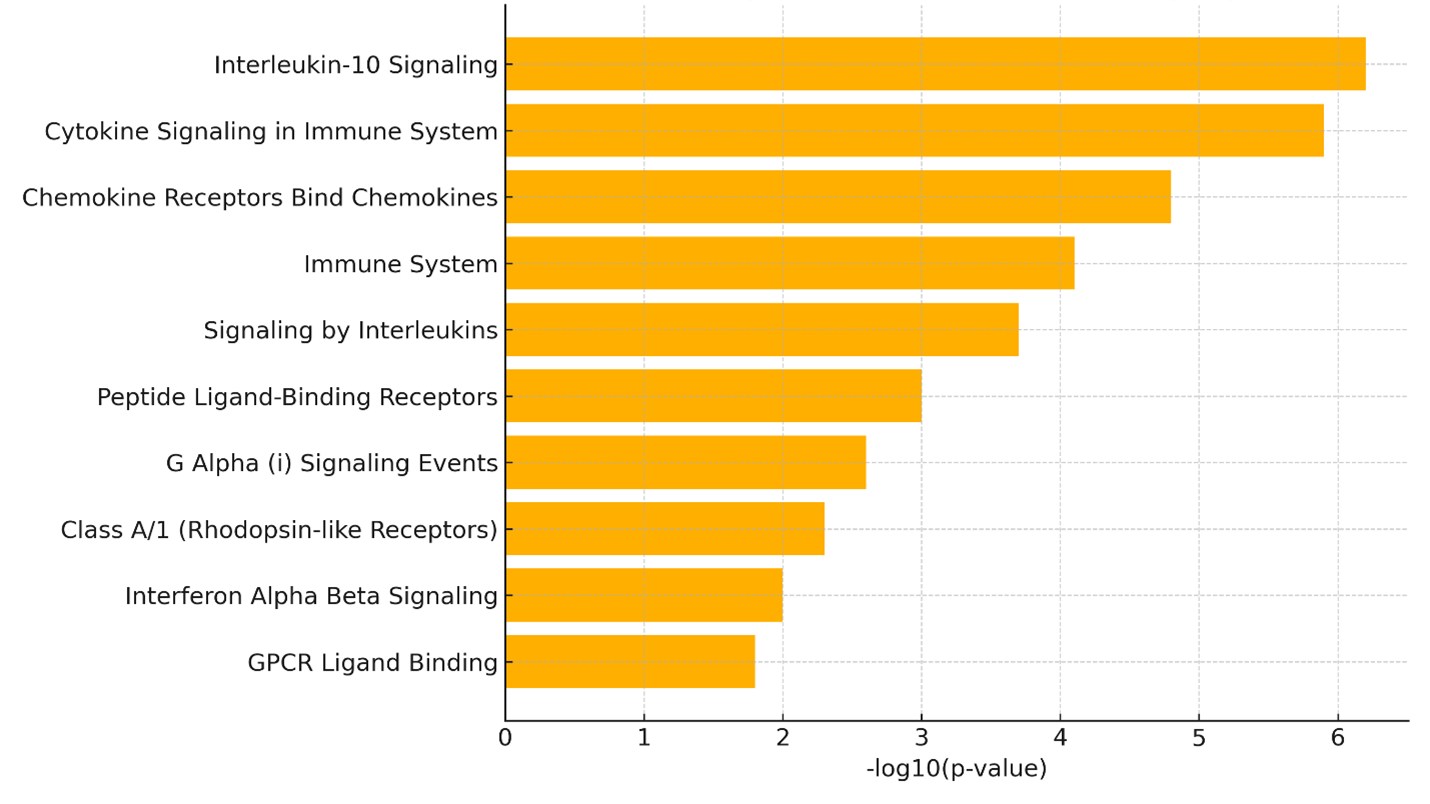
Reactome pathway enrichment analysis of genes upregulated in BDCA1+ lung dendritic cells. Bar plot displaying the top ten suggestively enriched Reactome pathways among the six most upregulated genes in BDCA1+ (CD1c+) dendritic cells (CCL5, CXCL8, ISG15, IFNB1, CXCL10, and IL1B). Bar lengths represent the –log10(p-value) of enrichment, with higher values indicating stronger statistical significance. Enriched pathways include Interleukin-10 signaling, Cytokine signaling in immune system, and Chemokine receptor interactions, underscoring the inflammatory and antiviral transcriptional programming of BDCA1+ DCs under baseline conditions. Notably, literature-supported regulators IRF7 and BATF3 were identified as potential upstream nodes linking interferon and antigen-presentation pathways.
Comparative Analysis with Single-Cell RNA-Seq Atlases
For external validation, we interrogated publicly available single-cell RNA-seq atlases—the Human Lung Cell Atlas (HLCA) and PanglaoDB—to compare immune-gene expression in BDCA1 (CD1c) and BDCA3 (CD141) lung dendritic cells. Both datasets reproduced the subset-specific pattern observed in the microarray analysis: BDCA1 cells displayed heightened basal expression of the pro-inflammatory mediators IL1B, CXCL8 and CCL5, whereas BDCA3 cells expressed higher levels of selected interferon-stimulated genes, notably ISG15 and IFIT1.
Quantitative comparison of mean single-cell expression further substantiated this divergence (
Comparative single-cell RNA-seq expression of selected immune genes in human lung dendritic cell subsets
| Gene | BDCA1+ Mean Expression | BDCA3+ Mean Expression | log2 fold change (BDCA1+ vs BDCA3+) |
|---|---|---|---|
| IL1B | 2.4 | 0.8 | 1.58 |
| CXCL8 | 3.1 | 1.2 | 1.37 |
| CCL5 | 2.8 | 0.9 | 1.64 |
| ISG15 | 1.2 | 2.5 | -1.06 |
| IFIT1 | 1 | 2.8 | -1.48 |
Collectively, this cross-platform concordance reinforces the robustness of our observations and underscores conserved transcriptional programmes that distinguish the two principal human lung dendritic-cell subsets.
Stimulus-Based Comparison from Prior Studies
To place the basal transcriptional divergence in context, we interrogated published datasets describing dendritic-cell responses to immune stimuli. In studies employing polyinosinic:polycytidylic acid (poly I:C), a synthetic double-stranded RNA that activates TLR3 and mimics viral infection, both BDCA1 (CD1c) and BDCA3 (CD141) dendritic cells up-regulated canonical maturation markers (CD40, CD80, CD86) together with antiviral mediators such as IFNB1 and CXCL10 11,12. Nevertheless, BDCA3 cells exhibited more pronounced induction of ISGs—including ISG15, IFIT1 and MX1—highlighting their specialisation for antiviral immunity and type I interferon signalling 13,14,15.
Conversely, BDCA1 cells mount a vigorous pro-inflammatory transcriptional programme upon viral challenge. For instance, respiratory syncytial virus (RSV) infection robustly induces IL1B, IL6, IL8, TNFα and IL12 in BDCA1 dendritic cells 16,17. Under identical conditions, BDCA3 cells elicit a comparatively tempered cytokine response yet up-regulate IL10, IL12, CD86 and co-inhibitory ligands such as PD-L1 16,18. These contrasting patterns are summarised schematically in
Stimulus-dependent immune responses of human lung dendritic cell subsets based on published studies
| Stimulus | DC Subset | Upregulated Markers / Cytokines |
|---|---|---|
| Poly I:C | BDCA1+ & BDCA3+ | CD40, CD80, CD86, IFNB1, CXCL10 |
| RSV | BDCA1+ | IL-1β, IL-6, IL-8, TNF‑α, CXCL10, IL-12 |
| RSV | BDCA3+ | IL-8, IL-10, IL-12; CD86, PD-L1 |
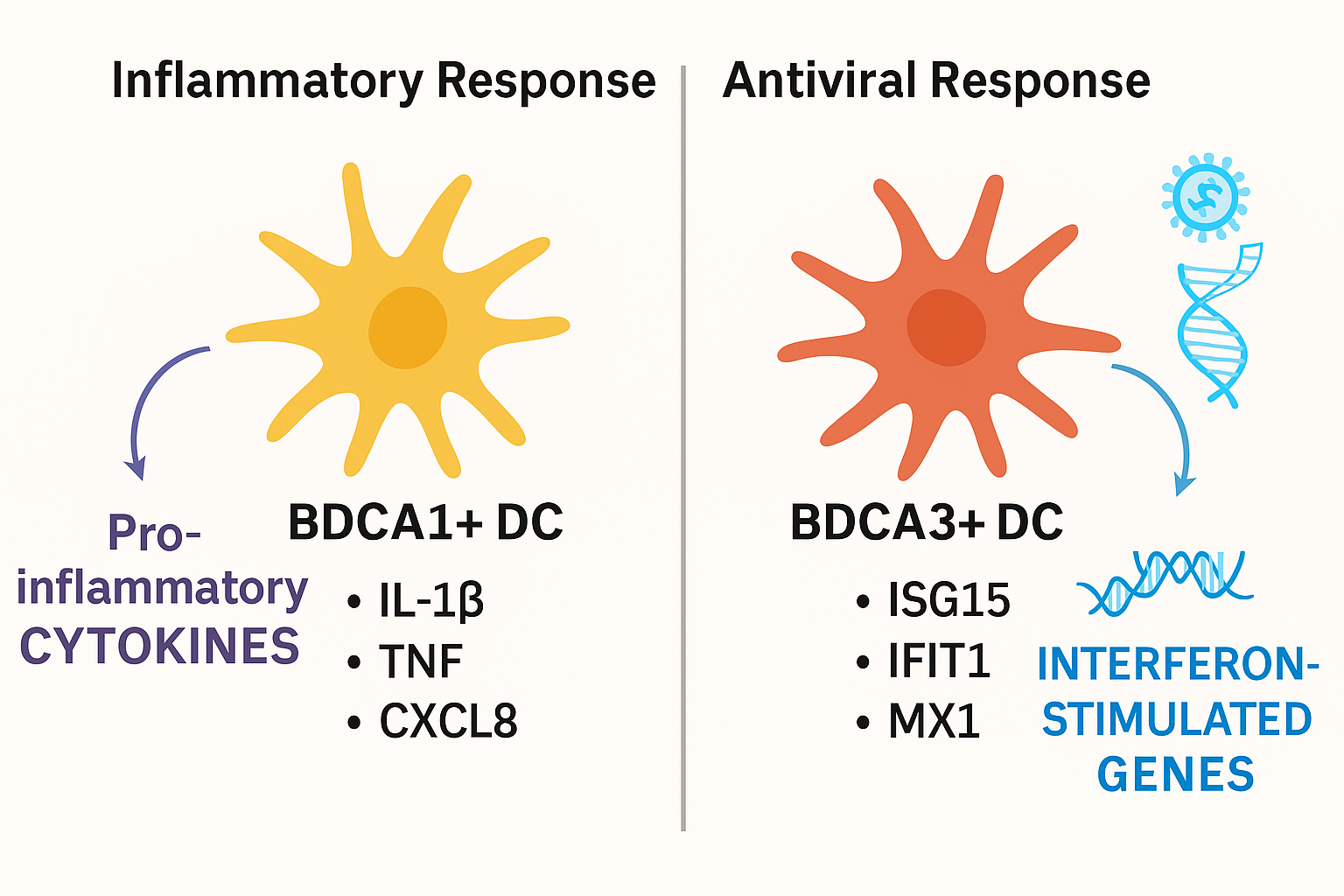
Functional divergence of BDCA1+ and BDCA3+ lung dendritic cells under baseline and stimulated conditions. Schematic illustration summarizing the distinct immunological roles of BDCA1+ (CD1c+) and BDCA3+ (CD141+) dendritic cells in the human lung. Under steady-state conditions, BDCA1+ DCs exhibit higher expression of pro-inflammatory cytokines and chemokines such as
Collectively, these comparative data indicate that the steady-state transcriptional dichotomy mirrors functional specialisation during pathogen encounter. BDCA1 cells are primed for rapid production of inflammatory cytokines and recruitment of effector leukocytes, whereas BDCA3 cells deploy a more regulated, interferon-driven antiviral programme. This functional separation affirms the biological relevance of our observations and underscores the necessity of subset-specific context in pulmonary immunity.
Exploratory Identification of Potential Regulatory “Switch” Factors
To further interpret the subset-specific expression landscape, we surveyed known transcriptional hierarchies associated with the leading differentially expressed cytokine- and interferon-related genes. Given their established roles and the ISG-biased profile of BDCA3 cells, the transcription factors IRF7 and BATF3—both pivotal for type I interferon signalling and antigen cross-presentation—emerge as plausible molecular ‘switches’ that delineate BDCA3 antiviral specialisation from BDCA1 inflammatory priming. IRF7 serves as a master activator of ISGs, including ISG15 and IFIT1, whereas BATF3 is indispensable for the development and function of cDC1 (BDCA3) cells. Collectively, these literature-backed links imply that divergent regulatory hierarchies, rather than stochastic variations, underpin the functional polarisation of human lung dendritic-cell subsets 20.
Discussion
In this study, we sought to validate and integrate previously reported functional distinctions between BDCA1 and BDCA3 dendritic cells (DCs) in the human lung by re-analyzing publicly available transcriptomic datasets and placing these findings within the context of recent single-cell and functional studies. Using microarray data from GSE43184, we observed distinct immune-gene expression profiles: BDCA1 DCs exhibited elevated basal levels of pro-inflammatory cytokines and chemokines, whereas BDCA3 DCs displayed comparatively higher expression of interferon-stimulated genes (ISGs). These observations corroborate and extend prior reports, positioning our analysis as a cross-validation of existing single-cell and bulk transcriptomic findings rather than a de novo discovery effort. The observed patterns closely mirror those reported by the Human Lung Cell Atlas, which identified similar subset-specific expression signatures using single-cell RNA-seq 3,6.
Our baseline analysis revealed that BDCA1 DCs express IL1B, CXCL8, CXCL10, and related inflammatory mediators at higher levels than BDCA3 DCs. This pattern indicates that BDCA1 DCs are transcriptionally primed for innate immune activation and amplification of inflammation, in agreement with their documented roles in T-helper-cell priming and recruitment of effector leukocytes 4,7. In contrast, BDCA3 DCs were enriched for ISGs such as ISG15 and IFIT1, supporting their established specialization in antiviral signaling and antigen cross-presentation 4.
Multivariate principal-component and hierarchical clustering analyses further reinforced the transcriptional separation of the two DC subsets. Reactome-based pathway enrichment revealed that BDCA1 DCs engage inflammatory, cytokine, and GPCR-related pathways—functional attributes characteristic of innate sentinels primed to respond to environmental threats.
Comparative validation using external resources strengthened these observations. Single-cell analyses from HLCA and PanglaoDB confirmed that the pro-inflammatory and chemotactic genes we identified as elevated in BDCA1 DCs are also differentially expressed in vivo 3,6. This cross-platform concordance indicates that even a small-sample microarray dataset can reliably reflect the intrinsic transcriptional polarization of human lung DC subsets. Likewise, studies employing poly I:C and RSV stimulation have shown that BDCA1 DCs mount robust inflammatory responses, whereas BDCA3 DCs preferentially activate antiviral and regulatory programs 7,8. Together, these lines of evidence support a coherent model in which baseline gene-expression patterns reflect pre-configured immune specializations that manifest during pathogen encounter.
The distinct molecular predispositions of BDCA1 and BDCA3 DCs have practical implications for respiratory immunity and translational applications. BDCA1 DCs, with their inflammatory bias, may respond rapidly to bacterial or fungal stimuli by producing cytokines that recruit neutrophils and T cells 7. In contrast, BDCA3 DCs, enriched in interferon signaling and cross-presentation pathways, appear optimised for antiviral defense and cytotoxic T-cell priming 4. These complementary programs likely contribute to the lung’s ability to balance inflammation with antiviral protection.
Our integrative analysis further highlights IRF7 and BATF3 as candidate transcriptional regulators that may govern the balance between inflammatory and antiviral programming. Consistent with previous studies demonstrating IRF7-driven interferon amplification and BATF3-dependent cDC1 differentiation 21,22, these factors align with the expression patterns observed here, where BDCA3 DCs exhibit ISG-enriched antiviral profiles and BDCA1 DCs display inflammatory-cytokine dominance. Although our study does not directly test transcription-factor activity, this literature-anchored inference provides a mechanistic framework for future experimental validation and underscores the value of cross-dataset synthesis in refining existing models of DC-subset specialization.
From a therapeutic perspective, selective modulation of DC subsets could inform vaccine and immunotherapy strategies. For instance, engaging BDCA3 DCs may enhance antiviral immunity through adjuvants that promote cross-presentation 10, while dampening BDCA1 DC-driven cytokine cascades could mitigate hyperinflammatory conditions such as acute respiratory distress syndrome (ARDS) or severe viral pneumonia (e.g., COVID-19) 11.
While our analysis offers integrative validation, several limitations warrant consideration. The small sample size (n = 2 per subset) in GSE43184 limits statistical power, and results should thus be viewed as supportive, confirmatory evidence rather than definitive proof. The microarray platform’s restricted gene coverage further limits discovery of novel transcripts and isoforms. Additionally, our inferences regarding responses to viral stimuli are based on secondary data rather than experimental validation. Future studies employing RNA-seq or functional assays could directly test these hypotheses. Despite these constraints, the reproducibility of expression trends across independent datasets and analytical platforms underscores the robustness of the observed subset specialization.
Conclusion
In summary, this validation-driven transcriptomic meta-analysis demonstrates that BDCA1 and BDCA3 dendritic cells (DCs) in the human lung are transcriptionally and functionally distinct, even at rest. BDCA1 DCs harbor a gene-expression signature enriched for inflammatory mediators, suggesting a central role in initiating and amplifying local immune responses. Conversely, BDCA3 DCs exhibit higher basal expression of interferon-stimulated genes, in line with their specialized antiviral and cross-presenting functions. Rather than revealing novel mechanisms, these data consolidate and extend previously reported differences via rigorous cross-dataset validation. The transcriptional divergence observed under steady-state conditions parallels the functional dichotomy seen after activation, underscoring the biological relevance of these baseline programs. Collectively, the study confirms that human lung DC subsets maintain stable, subset-specific transcriptional landscapes congruent with their established immunological roles, providing a framework for vaccine development, immunotherapeutic design, and targeted modulation of pulmonary inflammation.
Abbreviations
ARDS: Acute Respiratory Distress Syndrome; BATF3: Basic Leucine Zipper ATF-Like Transcription Factor 3; BDCA1: Blood Dendritic Cell Antigen 1; BDCA3: Blood Dendritic Cell Antigen 3; cDC1: Conventional Dendritic Cell Type 1; cDC2: Conventional Dendritic Cell Type 2; CD: Cluster of Differentiation; DCs: Dendritic Cells; FDR: False Discovery Rate; GEO: Gene Expression Omnibus; GPCR: G-Protein-Coupled Receptor; HLCA: Human Lung Cell Atlas; IFNB1: Interferon Beta 1; IL: Interleukin; IRF7: Interferon Regulatory Factor 7; ISGs: Interferon-Stimulated Genes; KEGG: Kyoto Encyclopedia of Genes and Genomes; PCA: Principal Component Analysis; PD-L1: Programmed Death-Ligand 1; poly I:C: Polyinosinic:Polycytidylic Acid; RMA: Robust Multi-array Average; RSV: Respiratory Syncytial Virus; scRNA-seq: Single-Cell RNA Sequencing; TLR: Toll-Like Receptor; TNF: Tumor Necrosis Factor
Acknowledgments
The author thanks the creators of the GSE43184 dataset for making their data publicly accessible through the Gene Expression Omnibus (GEO). All analyses were performed using open-source R and Python packages in a reproducible computational environment. The author also acknowledges the assistance of ChatGPT (OpenAI) for language editing and grammar refinement. The author read and approved the final manuscript and is solely responsible for the content and integrity of this work.
Author’s contributions
V.Y.: Conceptualization, Data Curation, Formal Analysis, Methodology, Visualization, Writing – Original Draft, Writing – Review & Editing. All authors read and approved the final manuscript.
Funding
This work did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors. The author declares that there are no financial conflicts of interest relevant to this article.
Availability of data and materials
All transcriptomic data used in this study are publicly available in the NCBI Gene Expression Omnibus (GEO) database. The primary microarray dataset analyzed (GSE43184) can be accessed at . External validation was performed using publicly available datasets from the Human Lung Cell Atlas and curated literature sources. All data processing, analysis scripts, and figure generation workflows are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Declaration of generative AI and AI-assisted technologies in the writing process
The authors declare that they have not used generative AI (a type of artificial intelligence technology that can produce various types of content including text, imagery, audio and synthetic data. Examples include ChatGPT, NovelAI, Jasper AI, Rytr AI, DALL-E, etc) and AI-assisted technologies in the writing process before submission.
Competing interests
The authors declare that they have no competing interests.

