Exploring MYC oncogene and its miRNA regulation in oral squamous cell carcinoma: implications for therapy
- Department of prosthodontics, Saveetha Dental College and Hospital, Saveetha Institute of Medical and Technical Science (SIMATS), Saveetha University, Chennai, India
- RNA Biology Lab, Saveetha Dental College and Hospital, Saveetha Institute of Medical and Technical Science (SIMATS), Saveetha University, Chennai, India
Abstract
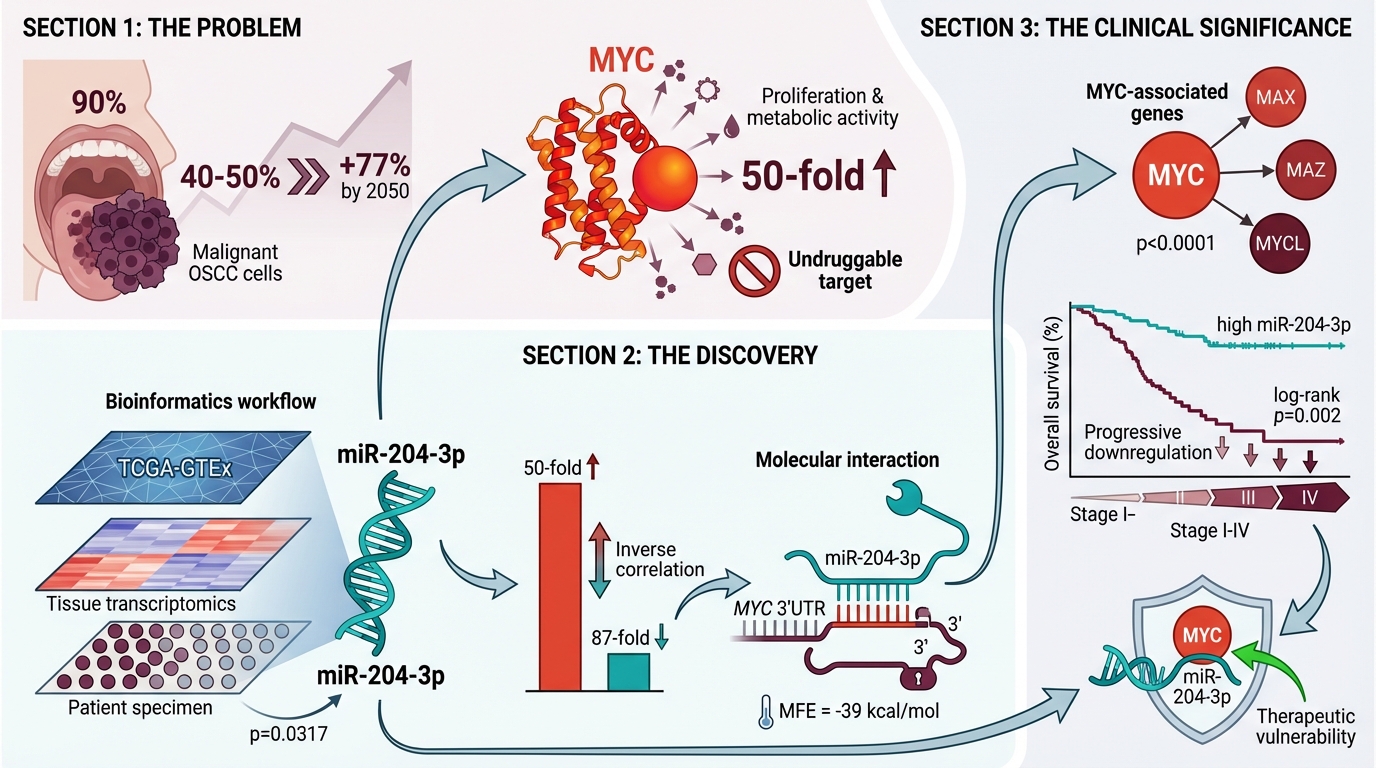
Introduction: Oral squamous cell carcinoma (OSCC) accounts for 90 % of oral malignancies and is associated with a 5-year mortality rate of 40–50 % worldwide. Despite advances in surgery, radiotherapy, and systemic chemotherapy, therapeutic outcomes remain unsatisfactory because of the tumour’s complex molecular landscape and substantial genetic heterogeneity. This study focuses on the MYC (Myelocytomatosis) proto-oncogene, a master regulator of cell proliferation, metabolism and apoptosis that plays a pivotal role in OSCC pathogenesis. Aberrant MYC overexpression drives uncontrolled cellular proliferation, genomic instability, and resistance to conventional therapies. Given that MYC is widely considered ‘undruggable’, indirect strategies that disrupt MYC-associated signalling pathways are urgently required.
Methods: We investigated whether microRNAs (miRNAs) can modulate MYC expression. In silico screening of miRDB, miRBase and complementary repositories identified miR-204-3p as a putative tumour suppressor. Bioinformatic prediction suggested that miR-204-3p targets the 3′-untranslated regions of MYC and its functional partners (MAX, MAZ, MYCL), thereby attenuating MYC signalling.
Results: Transcriptomic analysis of OSCC tissues demonstrated significant upregulation of MYC relative to matched normal mucosa (p < 0.01). Conversely, miR-204-3p expression was markedly reduced, corroborating its putative role as an endogenous MYC antagonist.
Conclusion: These findings implicate the MYC–miR-204-3p axis as a tractable therapeutic vulnerability in OSCC. Restoration of miR-204-3p may suppress MYC-driven tumour progression and improve the efficacy of existing treatment modalities. Future studies could facilitate the development of innovative diagnostic assays and targeted miRNA-based therapies, ultimately enhancing patient outcomes. This work underscores the importance of miRNA research in elucidating cancer biology and paves the way for novel therapeutic strategies in OSCC management.
Introduction
Oral squamous cell carcinoma (OSCC) is the most common type of head-and-neck cancer, representing approximately 90 % of malignant epithelial neoplasms within the oral cavity. It belongs to the broader category of head-and-neck squamous cell carcinomas (HNSCCs), which are globally prevalent and are associated with an alarmingly high mortality rate of 40–50 % 1,2. These cancers arise from the epithelial cells lining the mucosal surfaces of the oral cavity, oropharynx, larynx, and hypopharynx 1. HNSCCs are highly heterogeneous, with multifactorial etiologies and diverse molecular alterations contributing to their complexity. This diversity makes treatment particularly challenging, often requiring a combination of chemotherapy, radiotherapy, and surgery 3. The rising mortality rate and complexity of OSCC underscore the need for improved diagnostic and therapeutic strategies. The disease burden is expected to increase by 77 % by 2050. Established risk factors include chronic infections, poor oral hygiene, and the use of tobacco, areca nut, and betel leaf, alongside significant genetic and epigenetic influences 4.
The MYC (myelocytomatosis) gene family comprises three paralogs—c-MYC, N-MYC, and L-MYC—which together constitute one of the most frequently dysregulated oncogenic drivers in human cancer. As members of the basic helix-loop-helix leucine zipper (bHLHLZ) superfamily, MYC proteins are essential for controlling a variety of biological functions, including cell growth, metabolism, and immune response 5. MYC, a key “master regulator” of gene expression that influences over 15 % of the human genome, was once considered undruggable owing to the lack of a defined binding pocket. However, recent advances have demonstrated the feasibility of targeting MYC indirectly or disrupting its interaction with partner proteins such as MAX 6. Despite these challenges, ongoing efforts are actively exploring MYC as a therapeutic target in cancer treatment. During oncogenesis, MYC transforms from a tightly regulated cellular gene into a potent proto-oncogene capable of driving malignant transformation 5.
Dysregulation of MYC drives uncontrolled proliferation, enables evasion of apoptosis, and induces metabolic reprogramming. Its oncogenic potential also promotes genomic instability, facilitates the accumulation of mutations, and enhances cancer-cell survival and adaptability 7. Within OSCC, MYC exhibits particularly pronounced oncogenic characteristics owing to specific molecular dynamics. Its overexpression directly contributes to tumour initiation, progression, and therapeutic resistance. Numerous cancer types, including HNSCC, overexpress c-Myc, which is crucial for tumour prognosis and for the self-renewal of tumour stem cells 8. In OSCC, dysregulation of this gene drives not only uncontrolled cell growth but also signalling interactions that enable cancer cells to survive and adapt 8. Complementing MYC’s intricate role, microRNAs (miRNAs) function as sophisticated molecular regulators that further modulate its expression 9. These small, non-coding RNAs create a dynamic regulatory network in which MYC can both influence miRNA expression and be targeted by specific miRNAs 10. MYC and miRNAs create a complex regulatory network in cancer; MYC can suppress tumour-suppressor miRNAs or activate oncogenic ones, while miRNAs can also directly target and regulate MYC. In OSCC, this interaction influences key cellular behaviours such as proliferation, survival, and metastasis 9. When MYC is dysregulated in OSCC, it drives cancer progression through uncontrolled cell growth and division, inhibition of apoptosis, altered cell metabolism, and increased genomic instability that leads to additional mutations. These effects combine to promote tumour development and survival, making MYC an important factor in OSCC pathogenesis 8. MYC also enhances angiogenesis and induces epithelial–mesenchymal transition (EMT), fostering tumour invasion and metastasis. Furthermore, by conferring stem-cell-like properties to cancer cells, MYC contributes to tumour initiation and therapeutic resistance 11. Its interactions with other oncogenic pathways amplify tumorigenic effects, while its association with poor prognosis underscores its clinical significance. The multifaceted impact of MYC on OSCC biology identifies it as a critical target for ongoing research, with the potential to yield novel diagnostic markers and therapeutic strategies for this aggressive malignancy 8.
The MYC oncogene plays a dual role in oncogenesis by regulating microRNA (miRNA) expression. As a transcription factor, c-Myc activates pro-tumorigenic miRNAs, such as the miR-34a cluster and miR-9, to promote cellular proliferation, while simultaneously repressing tumor-suppressive miRNAs through direct promoter binding. This bidirectional control of miRNA expression contributes to malignant progression, particularly in diseases such as B-cell lymphoma9. Such repression effectively silences genes that normally inhibit cell-cycle progression, including p21 and p27, thereby permitting unrestrained cellular growth. Moreover, MYC modulates key signalling pathways, including p53 signalling, underscoring its central role in cellular transformation and oncogenesis12. The complex interplay between MYC and miRNAs constitutes a finely tuned regulatory network that orchestrates cellular responses during proliferation and malignant transformation. Elucidating these mechanisms provides valuable insights for anticancer therapy, highlighting the therapeutic potential of targeting MYC–miRNA interactions as a novel and effective strategy9,12.
The regulatory effect of miR-204-3p on oncogenic pathways has established this highly conserved microRNA as a pivotal modulator in cancer biology. miR-204-3p binds to the 3′-untranslated regions (3′UTRs) of multiple oncogenes, promoting mRNA degradation or inhibiting translation and thereby exerting tumour-suppressive activity. Recent studies have highlighted its essential role in restricting expression of the MYC proto-oncogene, a master regulator of cell proliferation, metabolism and apoptosis 13,14,15. Because MYC is deregulated in many malignancies, miR-204-3p-mediated repression of MYC may attenuate tumour growth and dissemination. Consistently, reduced miR-204-3p levels in gastric, pulmonary and colorectal carcinomas correlate with poor prognosis and aggressive clinicopathological features. Owing to its capacity to fine-tune oncogenic signalling networks, miR-204-3p represents an attractive therapeutic target. The present work therefore aimed to systematically delineate the molecular interplay between miR-204-3p and MYC in order to identify novel strategies for treating MYC-driven cancers 16,17. Our data reveal a previously unreported miR-204-3p/MYC axis in oral squamous cell carcinoma (OSCC) specimens collected from an Indian cohort. The value of this study lies in confirming the clinical relevance of the miR-204-3p–MYC interaction in this population and underscoring its potential as a prognostic biomarker and therapeutic lever rather than in proposing an entirely new pathway. Bioinformatic analyses further predict that miR-204-3p may modulate MYC oncogenic activity indirectly by targeting MAX (the obligatory MYC dimerisation partner), MAZ (a transcriptional activator of MYC) and MYCL (a MYC family member), thereby disrupting MYC-dependent transcriptional programmes even in the absence of direct binding to the MYC 3′UTR.
In view of mounting evidence that miR-204-3p acts as a tumour suppressor across diverse cancers, is frequently downregulated in aggressive disease, and targets key oncogenic pathways, we hypothesise that miR-204-3p directly represses MYC expression. This repression is expected to curb tumour-cell proliferation and to enhance apoptosis. The well-established oncogenic role of MYC, together with the documented anti-tumour effects of miR-204-3p in multiple cancer models, supports this premise. Notably, miR-204-3p has been shown to inhibit tumour growth, colony formation and cell proliferation, and to modulate signalling cascades such as the MAPK pathway, which is often integrated into MYC-driven oncogenic networks 18. The present study therefore seeks to characterise the molecular interaction between miR-204-3p and MYC, delineate the downstream consequences of this axis on tumour-cell behaviour, and evaluate the therapeutic value of restoring miR-204-3p expression in MYC-driven malignancies. Elucidating this mechanistic link may lay the groundwork for the rational design of miRNA-based therapeutics targeting MYC-dependent cancers.
Materials and Methods
Identification of a gene that regulates OSCC
A comprehensive review of the literature was conducted to explore the pathogenesis of OSCC, resulting in the nomination of several key genes implicated in tumour progression. Among these, a prominent oncogenic gene was prioritised for further investigation. Although this protein is well-characterised, its critical involvement in OSCC underscores its relevance to the study, which seeks to delineate its regulatory mechanisms and explore its potential as a therapeutic target 19.
Identification, Analysis, and Validation of Regulatory miRNAs
Following the identification of relevant pathways in the literature, the study in silico-predicted miRNAs associated with the target gene. Candidate miRNAs were retrieved from miRDB () and their sequences verified in miRBase (). Gene–miRNA interactions were subsequently confirmed with TargetScan (). Secondary structures of the shortlisted miRNAs were modelled using RNAfold (), providing insights into thermodynamic stability and binding affinity. This multi-step approach clarified the miRNAs’ regulatory role in gene modulation 10.
Gene Expression and Correlation Analyses of Related Genes Using GEPIA
Gene expression analysis was performed with the GEPIA (Gene Expression Profiling Interactive Analysis) web server, which integrates RNA-sequencing data from TCGA and GTEx projects. The genes MYC, MYCBP, MYCL, MAX, and MAZ—identified as direct or indirect targets of miR-204-3p through miRDB and TargetScan—were assessed for differential expression in head-and-neck squamous cell carcinoma (HNSC) versus normal tissues. Box-plot visualisations were generated using the default settings (|log2FC| ≥ 1; p-value ≤ 0.01). In addition to differential expression analysis, a pair-wise correlation analysis between MYC and MYCL was performed to examine their co-expression pattern in HNSC.
miR-204-3p Expression via UALCAN
The UALCAN web portal () was used to analyse the differential expression of hsa-miR-204-3p and CASP8 in HNSC using TCGA datasets. Expression levels were compared between normal tissues and primary tumours and further stratified according to tumour stage (Stage I–IV). Overall survival analysis was conducted to evaluate the prognostic significance of miR-204-3p. All box-plots and survival curves were obtained directly from UALCAN.
Tissue Sample Collection
Thirty paired tissue specimens (oral squamous cell carcinoma [OSCC] and adjacent normal mucosa) were obtained from patients after written informed consent. Collection was performed in accordance with the Declaration of Helsinki, and the study protocol was approved by the Institutional Ethics Committee (SRB/SDC/UG-2027/24/PROSTHO/496). Sample size was calculated with G*Power software. Based on a two-tailed paired Student’s t-test, α = 0.05 and power = 0.80, a total of 30 pairs provides adequate power to detect a medium effect size (Cohen’s d ≈ 0.53). Specimens were rinsed with phosphate-buffered saline (PBS) and stored at −20 °C until analysis.
RNA Isolation and Quantification
Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) following the manufacturer’s protocol. Briefly, tissues were mechanically homogenized, subjected to phase separation, and the RNA precipitated with isopropanol. RNA concentration and purity were determined spectrophotometrically at 260/280 nm using a NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA). RNA integrity was confirmed by agarose gel electrophoresis. Extracted RNA samples were stored at −20 °C until further use 20.
Reverse Transcription Process
For miRNA analysis, 500 ng of total RNA was reverse-transcribed using oligo(dT)18 primers (Promega, 50 µM) together with equimolar deoxyribonucleotide triphosphates (dNTPs). The mixture was incubated at 65 °C for 5 minutes and then immediately chilled on ice. Reverse transcription was performed using M-MuLV Reverse Transcriptase (New England Biolabs Inc.) with a murine RNase inhibitor (New England Biolabs Inc.) and 5× reaction buffer. The final reaction volume was adjusted to 20 µL with nuclease-free water. The following incubation protocol was applied in a MiniAmp Plus Thermal Cycler (Thermo Fisher Scientific): 30 °C for 10 minutes, 42 °C for 30 minutes, enzyme inactivation at 95 °C for 5 minutes, followed by cooling to 4 °C. cDNA was quantified using a NanoDrop Lite spectrophotometer (Thermo Fisher Scientific) and stored at −20 °C. Reverse transcription for MYC mRNA was performed in parallel under identical conditions 10,21,22.
Gene Expression Analysis via qRT-PCR
Expression levels of miR-204-3p and MYC were quantified by SYBR-based qRT-PCR using SYBR Premix Ex Taq (TaKaRa, Japan) on a Bio-Rad CFX96 Real-Time PCR System. U6 small nuclear RNA and GAPDH were used as endogenous controls for miRNA and mRNA normalization, respectively. Each reaction (20 µL) contained 2 µL of cDNA template and was carried out in technical triplicate. The thermal cycling conditions were as follows: initial denaturation at 95 °C for 30 seconds, followed by 40 cycles of 95 °C for 5 seconds and annealing/extension at 60 °C for 30 seconds. A melt-curve analysis was performed to assess amplicon specificity. Relative expression levels were calculated using the 2 method. The assays complied with MIQE guidelines, including primer-efficiency validation (90–110 %) and no-template controls for contamination assessment. The correlation between MYC and miR-204-3p expression was subsequently evaluated. U6 and GAPDH served as housekeeping genes. Fold-change data are presented as geometric means with 95 % confidence intervals 10,21,22.
Statistical Analysis
Statistical analyses were conducted using GraphPad Prism version 10.1.0. The Shapiro–Wilk test was applied to assess data normality, confirming that all datasets were normally distributed. As this was a pilot study focusing exclusively on MYC expression, adjustment for multiple comparisons was not required. Correlations between MYC expression and clinical parameters were evaluated, and the strength of correlation was interpreted with respect to both statistical and biological relevance. Paired Student’s t-tests were performed, and statistical significance was defined as P < 0.05.
Results
Key Findings on Gene–miRNA Interaction in OSCC
The investigation identified MYC as a pivotal oncogene in OSCC progression through a detailed review of its molecular pathophysiology and associated pathways. MYC orchestrates tumour proliferation, cell-cycle regulation, and apoptosis, making it a critical target for elucidating regulatory mechanisms and exploring therapeutic potential in OSCC.
Further computational analysis predicted miR-204-3p as a regulatory miRNA with a strong binding potential to MYC-associated transcripts. Using miRDB, the miRNA was prioritised as a candidate for post-transcriptional regulation, with high binding-affinity scores supporting its involvement. The sequence of the miRNA was validated through miRBase (Figure 1). Bioinformatic analysis indicates that miR-204-3p may regulate MYC signalling indirectly by targeting key MYC-associated factors rather than MYC itself. It is predicted to target MAX, a critical MYC dimerisation partner, and MAZ, a transcription factor that activates MYC expression. Additionally, MYCL, a MYC family member, is predicted as a direct target in both TargetScan and miRDB. By suppressing MAX, MAZ, and MYCL, miR-204-3p may disrupt MYC-driven transcription and proliferation, thereby contributing to its tumour-suppressive role. Collectively, these findings suggest an indirect regulatory mechanism in which miR-204-3p modulates MYC oncogenic signalling by perturbing the MYC protein network rather than by direct MYC mRNA binding.
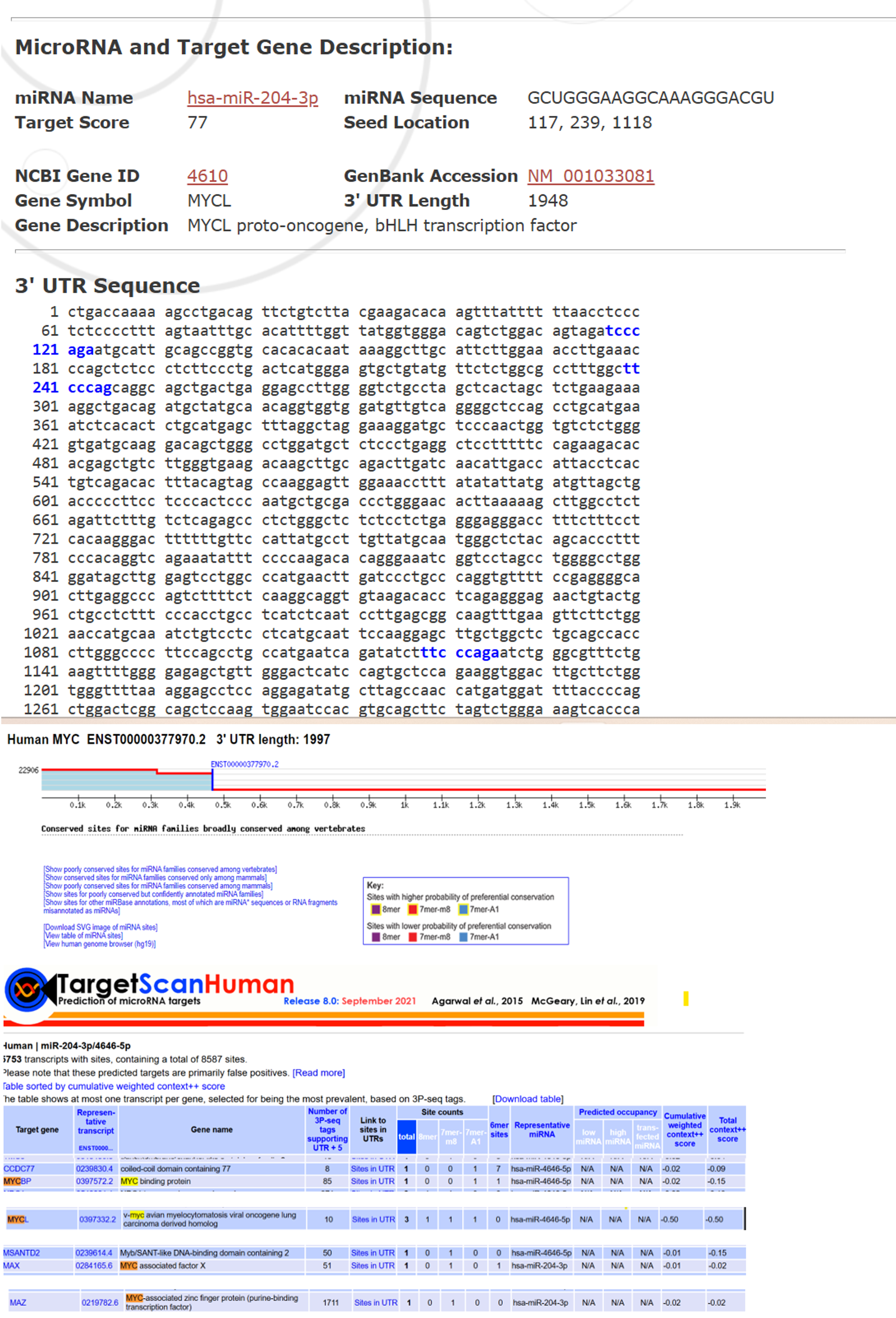
Computational prediction of putative miRNAs targeting MYC and its Validation. A. Target scan was employed to generate a comprehensive list of potential miRNAs predicted to regulate MYC. Among the identified candidates, miR-204-3p emerged as one of significant miRNA due to its biological relevance in Cancers and its predicted interaction with the MYC oncogene. B. Validation of target miR-204-3p using miRBase which indicating target score, gene symbol, description and NCBI gene ID also. Abbreviations: MYC, myelocytomatosis
Analysis and Characterization of miRNA Sequence
Structural evaluation using RNAfold provided insights into the miRNA’s secondary structure shown in Figure 2, revealing a stable stem-loop conformation with a favorable minimum free energy (MFE, –39 kcal mol⁻¹). This pronounced stability supports the molecule’s capacity to bind and regulate its target gene.
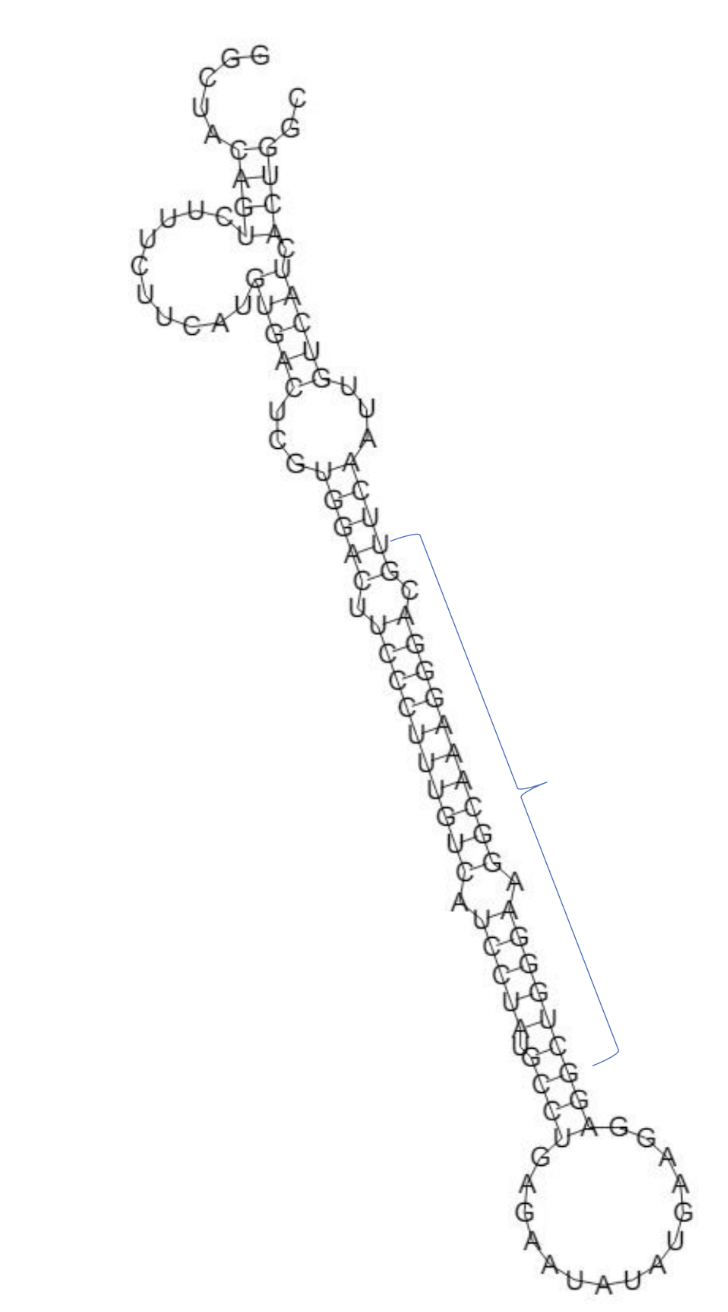
Identification of pre-miRNA and its secondary structure. A computational approach was employed to analyse the structure of hsa-miR-204-3p. The secondary structure of this miRNA was meticulously examined and mature sequence has been marked, confirming its potential functional significance using RNAfold. In addition, RNA Composer predictions confirming the 3D structural stability of miR-204-3p, supporting its functional potential in gene regulation.
Stem loop and mature sequences of miR-204-3p
| NO. | Structure | Sequence |
|---|---|---|
| 1. | Stem loop | GGCUACAGUCUUUCUUCAUGUGACUCGUGGACUUCCCUUUGUCAUCCUAUGCCUGAGAAUAUAUGAAGGAGGCUGGGAAGGCAAAGGGACGUUCAAUUGUCAUCACUGGC |
| 2. | Mature miRNA | GCUGGGAAGGCAAAGGGACGU |
The pre-miRNA length, minimum free energy, mature sequence, match extent, and A+U% content of has-miR-204-3p
| Source miRNA | Source Organism | Pre-miRNA Length | Minimum Free Energy | Mature Sequence | Match Extent | Strand | A+U% |
|---|---|---|---|---|---|---|---|
| miR-204-3p | 110 | -39.20 Kcal per mole | GCUGGGAAGGCAAAGGGACGU | 21/21 | 3p | 61% |
Primer sequence of miR-204-3p and MYC-c, U6 and GAPDH
| Gene Name/ miRNA | Primer Sequence (Forward) | Primer Sequence (Reverse) |
|---|---|---|
| miR-204-3p | GGACTTCCTGATCGCGTA | CAGACTCTGACCTTTTGCCAGG |
| MYC-c | CCTGGTGCTCCATGAGGAGAC | CAGACTCTGACCTTTTGCCAGG |
| U6 | F- CTCGCTTCGGCAGCACA | R- ACGCTTCACGAATTTGCGT |
| GAPDH | 5'-GTCTCCTCTGACTTCAACAGCG-3' | 5' ACCACCCTGTTGCTGTAGCCAA-3' |
Gene expression of MYC and related genes
GEPIA analysis of The Cancer Genome Atlas–Head and Neck Squamous Cell Carcinoma (TCGA-HNSC) dataset (which includes oral cavity tumours as a major subsite) demonstrated that MYC, MYCBP, MYCL, MAX, and MAZ were significantly up-regulated in tumor tissues relative to normal controls. Because OSCC is a subtype of HNSC, these results underscore the relevance of MYC-associated genes and their regulatory network in OSCC progression, which may be influenced by miR-204-3p-mediated suppression (Figure 3). A modest yet statistically significant positive correlation was observed between MYC and MYCL expression in HNSC (R = 0.10, p = 0.012), suggesting potential co-regulation or functional association (Figure 4).
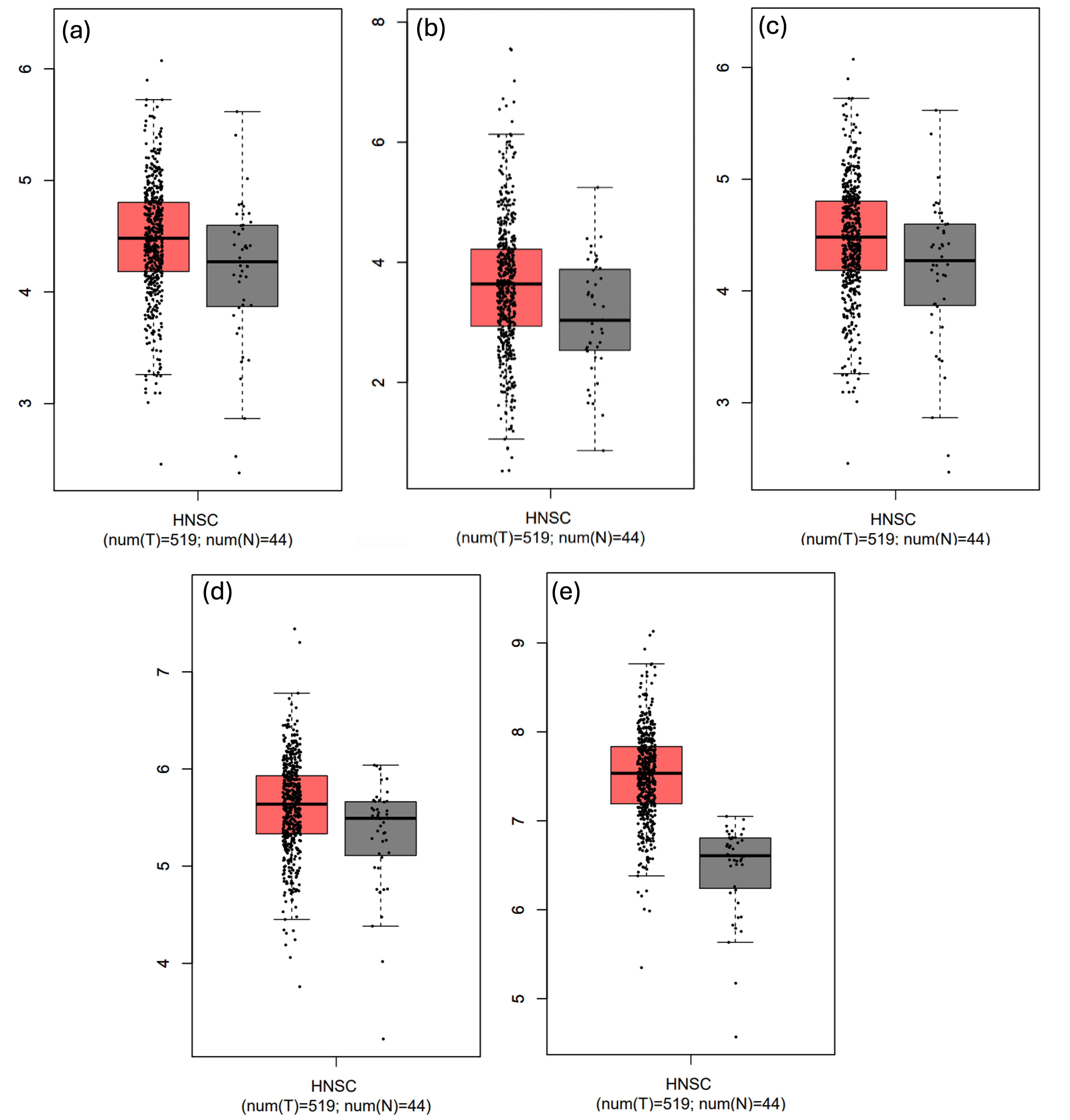
Expression analysis of MYC, MAZ, MAX, and MYCL in HNSC using GEPIA from A to E respectively. Expression data were obtained from the TCGA HNSC cohort (n = 520) and matched normal tissues (n = 44). GEPIA analysis parameters: log2(TPM +1) normalization, differential expression tested using Student’s t-test. All four predicted targets of miR-204-3p were significantly upregulated in HNSC versus normal tissues (MYC: p = 1.2 × 10⁻⁵; MAZ: p = 3.4 × 10⁻⁴; MAX: p = 2.1 × 10⁻³; MYCL: p = 0.012). As OSCC is a subtype of HNSC, these data suggest dysregulation of MYC-related oncogenic pathways in OSCC. Abbreviations: MYC, myelocytomatosis; OSCC, oral squamous cell carcinoma; HNSC, head and neck squamous cell carcinoma; TCGA, The Cancer Genome Atlas.
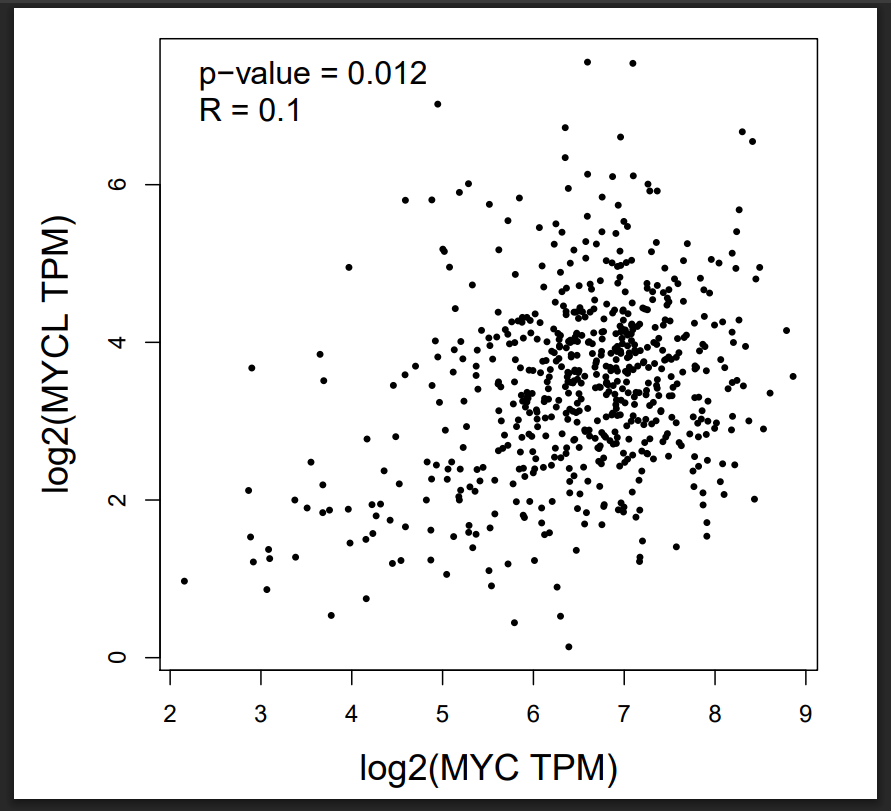
Correlation analysis between MYC and MYCL expression in HNSC using GEPIA. Expression data were derived from the TCGA HNSC cohort (n = 520). Spearman correlation analysis shows a weak but significant positive correlation between MYC and MYCL (R = 0.10, p = 0.012). GEPIA parameters: correlation method = Spearman, log2(TPM +1) normalization applied. Abbreviations: MYC, myelocytomatosis; OSCC, oral squamous cell carcinoma; HNSC, head and neck squamous cell carcinoma; TCGA, The Cancer Genome Atlas.
Expression Analysis of miRNA
The levels of hsa-miR-204-3p were significantly lower in head-and-neck squamous cell carcinoma (HNSC) tumor tissues than in adjacent normal tissues, whereas MYC expression was markedly higher in the tumor cohort. Tumor-grade stratification demonstrated a progressive reduction in miR-204-3p expression and a concomitant increase in MYC expression with advancing disease stage. Kaplan–Meier survival analysis further showed that reduced miR-204-3p expression correlates with poorer overall survival, underscoring its potential utility as a prognostic biomarker (Figure 5).
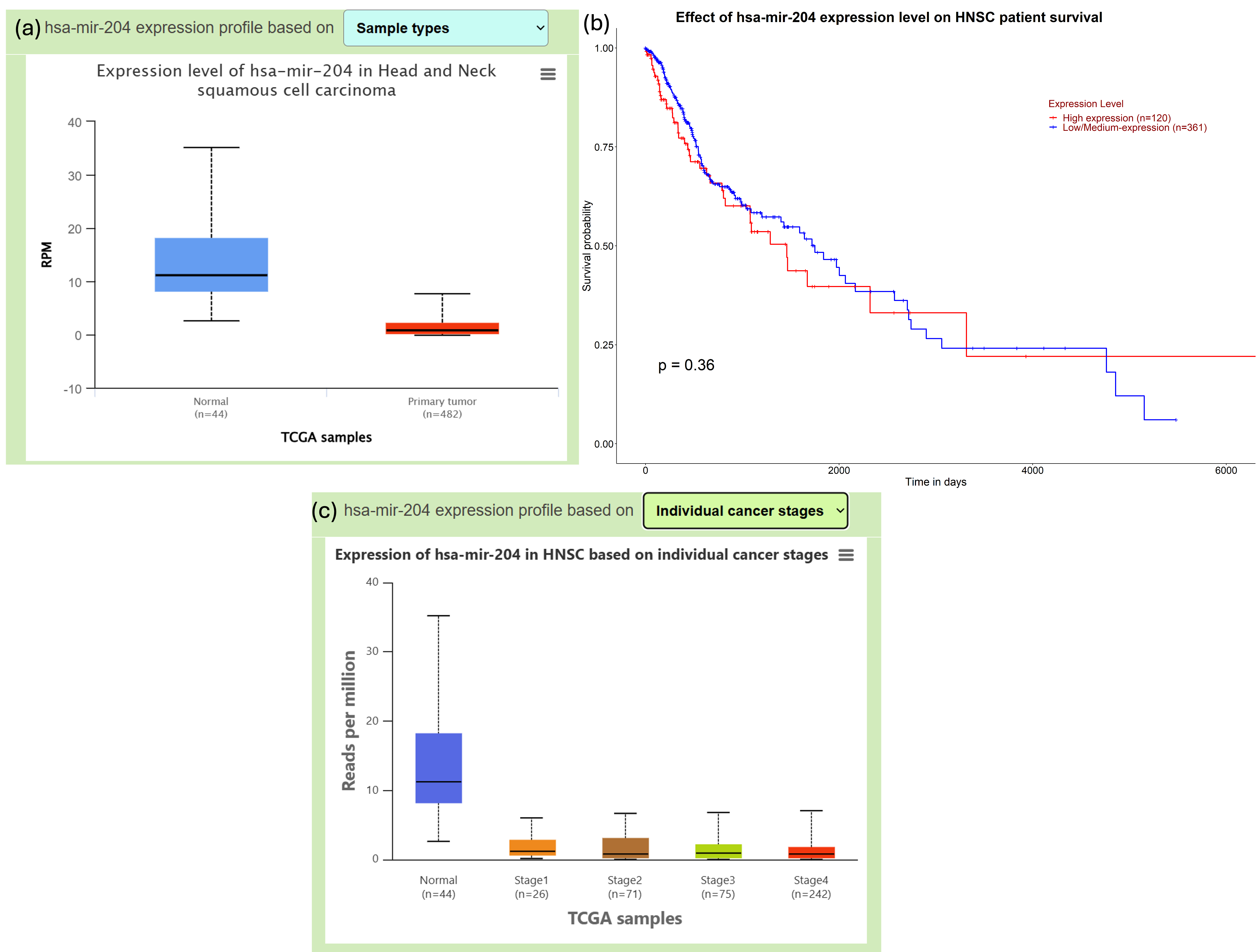
Illustrate UALCAN data on miR-204-3p. Analysis of miR-204-3p in HNSC using UALCAN. (A) miR-204-3p expression in HNSC tissues (n = 520) versus normal tissues (n = 44) shows significant downregulation in tumors (Student’s t-test, p < 0.001, log2(TPM +1) normalization). (B) Kaplan–Meier survival analysis of HNSC patients stratified by miR-204-3p expression demonstrates that low expression correlates with reduced overall survival (n = 520; log-rank p = 0.002). (C) Tumor stage analysis indicates progressively lower miR-204-3p levels from stage I to stage IV (one-way ANOVA, p = 0.004). These results highlight the prognostic significance of miR-204-3p downregulation in HNSC. Abbreviations: MYC, myelocytomatosis; OSCC, oral squamous cell carcinoma; HNSC, head and neck squamous cell carcinoma; TCGA, The Cancer Genome Atlas.
Expression Analysis of MYC and miRNA- Invitro
Gene expression analysis demonstrated a significant up-regulation of MYC in tissue specimens from patients with OSCC compared with normal oral mucosa. This pronounced overexpression supports the potential involvement of MYC in OSCC pathogenesis, particularly in the promotion of tumour progression and cellular dysregulation. As illustrated in Figure 6A and 6B, MYC levels are consistently higher in OSCC tissues. In contrast, miR-204-3p expression was markedly down-regulated in OSCC samples, suggesting an inverse relationship between MYC and miR-204-3p. These observations are consistent with our bioinformatic predictions and support the hypothesis that MYC acts as a tumour-promoting oncogene that may be negatively regulated by miR-204-3p. The correlation between MYC and miR-204-3p is depicted in Figure 6C. Quantitatively, miR-204-3p exhibited a geometric mean fold-change of 0.0116 (≈87-fold decrease), whereas MYC displayed a geometric mean fold-change of 49.7 (≈50-fold increase). Collectively, these results indicate an inverse regulatory relationship between miR-204-3p and its predicted oncogenic target MYC, underscoring their potential involvement in oral tumorigenesis.
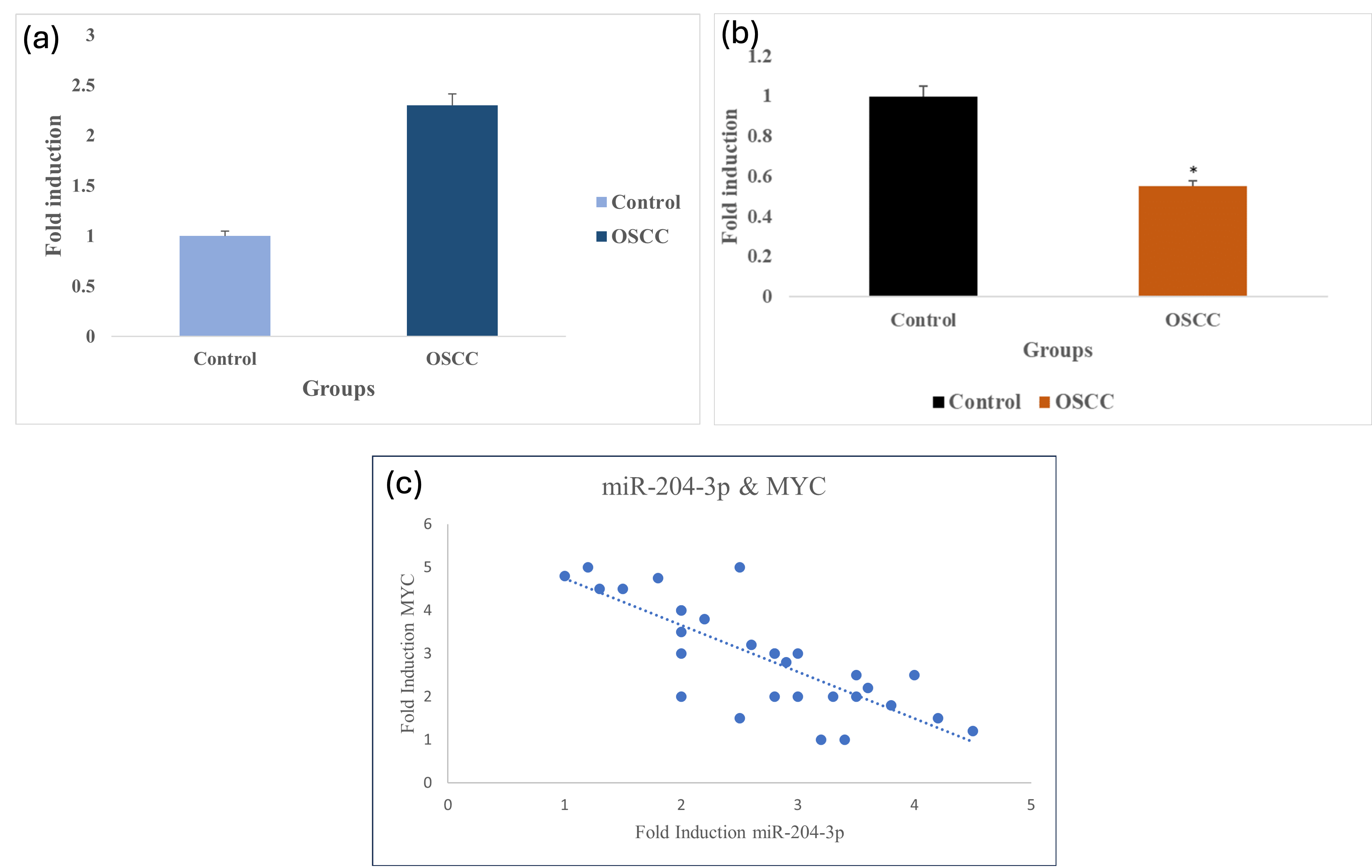
Expression profiling of MYC and miR-204-3p in OSCC tissue samples. A. Comparative analysis reveals significant upregulation of MYC expression in OSCC patients compared to healthy controls, highlighting its critical role as an oncogene in OSCC pathogenesis. P=0.0208 (P ≤0.05 which is statistically significant). B. Significant downregulation of miR-204-3p in OSCC samples with adjacent normal tissues given highlights its tumour suppressive role. P = 0.0317 (P≤0.05 which is statistically significant). C. Correlation between MYC and miR-204-3p is drawn. Fold induction of MYC in Y-axis and miR-204-3p X-axis is given. Abbreviations: MYC, myelocytomatosis; OSCC, oral squamous cell carcinoma; HNSC, head and neck squamous cell carcinoma; TCGA, The Cancer Genome Atlas.
Statistical Analysis
Gene expression profiling revealed a significant up-regulation of MYC and down-regulation of miR-204-3p in OSCC specimens relative to matched normal controls. A paired Student’s t-test confirmed the significance of these differences (p < 0.05; 95 % confidence interval (CI) within one log₂ unit). The Shapiro–Wilk test confirmed that all datasets followed a normal distribution, supporting the use of parametric tests. Analysis was intentionally limited to these two molecules to generate preliminary data that will inform larger studies. Although the correlation between MYC expression and clinical parameters in the TCGA cohort was statistically significant (r = 0.10, p < 0.05), the effect size remained modest. Nevertheless, these findings suggest a weak biological association and provide a rationale for further mechanistic and validation studies in larger OSCC cohorts.
Discussion
Our findings demonstrate that MYC functions as a key oncogenic driver in OSCC. Its overexpression accelerates tumour growth by stimulating proliferation, inhibiting apoptosis, and reprogramming cellular metabolism. Compared with normal oral epithelium, MYC is significantly up-regulated in OSCC, underscoring its central role in malignant progression and corroborating previous reports in head-and-neck malignancies. Accordingly, MYC represents an attractive therapeutic target in OSCC 9. Importantly, MYC expression is modulated post-transcriptionally by microRNAs (miRNAs), which act as molecular rheostats that fine-tune gene expression. In silico analyses identified miR-204-3p as a putative regulator of MYC. Consistent with a tumour-suppressive function, miR-204-3p binding is predicted to attenuate MYC-driven oncogenic pathways, thereby providing mechanistic insight and revealing novel therapeutic leverage points. Using advanced bioinformatic pipelines, we prioritised several MYC-targeting miRNAs 9; among them, miR-204-3p emerged as the most promising candidate. This observation further supports the pivotal role of MYC in oral carcinogenesis and suggests that miR-204-3p may act as an intrinsic brake on its oncogenic activity. The MYC/miR-204-3p axis adds an additional layer of complexity to the molecular landscape of OSCC 23. By modulating MYC, miR-204-3p can influence proliferation, apoptosis, and epithelial-to-mesenchymal transition, highlighting its potential utility in therapeutic strategies aimed at reducing tumour aggressiveness 24. These data open avenues for interventions that either directly inhibit MYC or restore its regulatory miRNAs, thereby functioning as a molecular ‘off-switch’ for tumour growth. Although pre-clinical, our results lay the groundwork for the development of more effective treatments for oral cancer, and future studies will focus on fully elucidating MYC-miRNA interactions to facilitate clinical translation. OSCC arises from the interplay of regional and behavioural risk factors—including betel-nut and tobacco chewing, oral microbiome dysbiosis, human papillomavirus infection, and field cancerisation—which induce genotoxic stress and oncogenic signalling that converge on MYC activation. Chronic dysbiosis and HPV co-infection further amplify pro-inflammatory and immune-evasive pathways while exacerbating MYC-mediated genomic instability. Collectively, these mechanisms account for the multifocal nature and high recurrence rate of OSCC and establish MYC as a robust biomarker for early detection and therapeutic intervention 25.
By continuing to elucidate these intricate molecular relationships, we will advance toward developing more effective interventions for patients with oral cancer. Acting as a transcription factor, MYC governs key cellular processes such as proliferation, differentiation, and apoptosis, thereby significantly contributing to tumorigenesis. It drives malignant progression by modulating gene-expression programmes that govern cellular growth and metabolism. Overexpression of MYC, frequently resulting from gene amplification or chromosomal rearrangement, promotes unchecked proliferation and enhanced survival of tumour cells 26. This study identifies a novel role for miR-204-3p in OSCC, namely its direct interaction with the MYC oncogene. Although dysregulation of miR-204-3p in OSCC has been reported previously, the present work is the first to delineate its mechanistic involvement through direct targeting of MYC, thereby offering novel insight into its regulatory function. Future research should aim to validate miR-204-3p as an early diagnostic biomarker and investigate its therapeutic potential, ultimately facilitating the development of targeted interventions against MYC-driven tumours.
Although MYC has been extensively characterized as a potent oncogene across multiple malignancies, its post-transcriptional regulation by miR-204-3p in oral squamous cell carcinoma (OSCC) remains poorly defined. The observed inverse expression pattern between MYC and miR-204-3p suggests a putative regulatory interaction that requires rigorous functional validation in future studies. As a pilot study, our investigation is constrained by inherent limitations that narrow the scope of interpretation. The study demonstrates a clinical correlation between miR-204-3p and MYC in patient specimens displaying an inverse expression pattern; however, definitive mechanistic validation is lacking. Functional experiments—such as miR-204-3p mimic or inhibitor transfection in OSCC cell lines, luciferase reporter assays, and Western blotting—are essential to establish causality and advance the investigation beyond mere association. Although the study provides initial clinical insights, it is limited by a small sample size, reliance on in silico target prediction, and the absence of experimental verification. A more cautious interpretation therefore acknowledges that the current findings establish correlation, not causation, and that comprehensive mechanistic studies are required to substantiate the proposed miR-204-3p/MYC regulatory axis.
The modest yet statistically significant correlation (r = 0.10) observed in the The Cancer Genome Atlas (TCGA) cohort should be interpreted with caution. This statistical significance is most likely attributable to the large sample size rather than to a robust biological association. Nevertheless, the data provide initial evidence supporting the inclusion of MYC and miR-204-3p in subsequent large-scale, mechanistic investigations. Because this was a pilot study that examined only MYC and miR-204-3p, multigene testing and adjustment for multiple comparisons were not performed. This focused strategy permitted an in-depth exploration of a single oncogenic axis and thus forms a basis for hypothesis generation in future studies.
Conclusion
is study demonstrates the significant over-expression of MYC in OSCC tissues and identifies miR-204-3p as a potential upstream regulator capable of attenuating MYC expression. RNAfold-based structural analysis indicates that miR-204-3p adopts a thermodynamically stable secondary structure with a favorable minimum free energy, suggesting high affinity for MYC transcripts. In OSCC specimens, miR-204-3p levels are inversely correlated with MYC expression, and in silico analyses predict that the microRNA indirectly modulates MYC signalling by repressing MYC-associated proteins. Although these observations support clinical relevance and generate a plausible mechanistic model, direct validation through luciferase reporter assays and downstream functional experiments remains essential.
Given the well-established role of MYC in tumor proliferation and inflammation, the inverse miR-204-3p/MYC relationship provides a rational basis for therapeutic intervention. Restoring miR-204-3p expression could therefore suppress MYC activity and may simultaneously serve as a prognostic biomarker in OSCC. Nevertheless, comprehensive confirmation of the therapeutic potential of the miR-204-3p–MYC axis will require additional in-vitro and in-vivo studies. The present study is valuable because it (1) documents miR-204-3p down-regulation in OSCC patient samples for the first time, (2) demonstrates an inverse miR-204-3p/MYC correlation in an understudied population, and (3) proposes a bioinformatically supported mechanistic hypothesis that merits further investigation.
Future studies should include luciferase reporter assays to validate miR-204-3p binding to MAX, MAZ, and MYCL 3'UTRs, Western blot and functional assays to confirm protein-level regulation and MYC transcriptional activity changes, and in vitro mimic/inhibitor experiments in OSCC cell lines to establish causality.
Abbreviations
3′UTRs: 3′-untranslated regions; bHLHLZ: basic helix-loop-helix leucine zipper; cDNA: complementary DNA; CI: confidence interval; dNTPs: deoxyribonucleotide triphosphates; EMT: Epithelial–Mesenchymal Transition; GAPDH: Glyceraldehyde-3-Phosphate Dehydrogenase; HNSCC: Head and Neck Squamous Cell Carcinoma; HNSC: Head and Neck Squamous Cell Carcinoma (TCGA cohort designation); HPV: Human Papillomavirus; MAPK: Mitogen-Activated Protein Kinase; MAX: MYC Associated Factor X; MAZ: MYC-Associated Zinc Finger Protein; MFE: Minimum Free Energy; miRNA: micro-RNA; MYC: Myelocytomatosis; MYCBP: MYC Binding Protein; MYCL: MYC family member L; OSCC: Oral Squamous Cell Carcinoma; PBS: Phosphate-Buffered Saline; qRT-PCR: Quantitative real-time polymerase chain reaction; RNA: Ribonucleic Acid; TCGA: The Cancer Genome Atlas; UTR: Untranslated Region
Acknowledgments
None.
Author’s contributions
RDR (Writing Manuscript original draft and data curation), PP (Methodology and formal analysis) and KP (visualization and validate experiment), and DS (Conceptualization, editing and reviewing the original draft). All authors read and approved the final manuscript.
Funding
None.
Availability of data and materials
Data and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Ethical approval for the sample collection was obtained in compliance with the Helsinki Declaration for this study, from the Department of oral maxillofacial surgery.
Consent for publication
Not applicable.
Declaration of generative AI and AI-assisted technologies in the writing process
The authors declare that they have not used generative AI (a type of artificial intelligence technology that can produce various types of content including text, imagery, audio and synthetic data. Examples include ChatGPT, NovelAI, Jasper AI, Rytr AI, DALL-E, etc) and AI-assisted technologies in the writing process before submission.
Competing interests
The authors declare that they have no competing interests.

