
Evaluation of P21Cip1 and P27Kip1 expression in de novo acute lymphoblastic leukemia patients
- Cancer prevention research center, Isfahan University of Medical Sciences, Isfahan, Iran
- HSCT Research Center, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- Laboratory Hematology and Blood Banking Department, School of Allied medical sciences, Shahid Beheshti University of Medical Sciences, Tehran, Iran
- Pediatric Congenital Hematologic Disorders Research center, Shahid Beheshti University of Medical Sciences, Iran
- Student Research Committee, Sabzevar University of Medical Sciences, Sabzevar, Iran
Abstract
Background: Acute lymphoblastic leukemia (ALL) arises from an imbalanced proliferation and differentiation of lymphoid progenitors due to special chromosomal and epigenetic abnormalities affecting cell cycle regulation. The cyclin-dependent kinase inhibitor (CDKI) family has crucial functions in G1 progression and G1 to S entry regulation. Among CDKIs, P21 and P27 are able to exert remarkable effects on all CDKs. Hence, we investigated the expression levels of P21 and P27 in ALL patients to determine whether or not their expression had been altered.
Materials and Methods: In the present study, we evaluated P21 and P27 expression in bone marrow and peripheral blood samples of 52 newly diagnosed ALL patients (30 males, 22 females) and 13 healthy normal controls (5 males, 8 females) using quantitative real-time PCR. Data were analyzed via SPSS (version 16) software and P<0.05 was assigned as the statistical significance level.
Results: Our findings demonstrated lower expression levels of P21 and P27 in ALL patients compared with normal controls (8.33- and 1.69-fold change, respectively). P21 and P27 expression was significantly different between T-cell acute lymphoblastic leukemia (T-ALL) and B-cell acute lymphoblastic leukemia (B-ALL) patients (P= 0.03).
Conclusion: Since P21 and P27 are able to influence the activity of both cyclins and CDKs, it is postulated that decreased expression of these genes reduces P21- and P27- mediated suppressive effects on cyclins and CDKs. Therefore, these events facilitate the activation of cyclins and CDKs which may result in cancer progression in ALL patients.
Background
Acute lymphoblastic leukemia (ALL) originates from blocked differentiation of lymphoid progenitors and affects both children and adults 1. Recent studies have tried to reveal the precise events contributing to the pathogenesis of ALL in order to enhance the effectiveness of therapeutic strategies in high-risk adult and childhood patients 2. Generally, ALL arises due to genetic lesions in genes involved in cell cycle regulation, apoptosis, and survival 3. Cell cycle regulators consist of families of cyclin-dependent kinase (CDK) inhibitors (CDKIs), including INK4 (p15, p16, p18, and p19) and CIP/KIP (P21CIP, P27KIP1, and p57KIP2) 4.
P21Cip1 and P27Kip1 are crucial CDK2 inhibitors which prevent G1 to S entry. The regulation of CDKI expression is considered as a common mechanism for cell integrity. In this regard, P21 or P27 over-expression acts as a vital component in tumor growth suppression. Furthermore, P21 and P27 are implicated in senescence and terminal differentiation of various types of cells, including hematopoietic cells. Several studies have demonstrated that P21 knockout mice are affected by different tumors, such as sarcomas and lymphomas. However, P21 gene mutations are infrequently detected in cancer cells. Moreover, decreased expression of P27 has been reported in a variety of cancers and contributes to tumor development 5. In primary T-ALL cells, P27 downregulation was associated with cell cycle progression and apoptosis inhibition 6.
Interestingly, recent studies have indicated a correlation between DNA methylation and decreased expression of some KIP family genes, such as P21, in ALL patients. Nevertheless, promoter methylation pattern of KIP family genes indicated inconsistent results. Traditionally, DNA damage induces cell cycle arrest via P21 and P27 activation. Although P21 and P27 have been considered as tumor suppressors in some studies, they have emerged as potential oncogenes in other studies as well. More studies have indicated that P21 and P27 could have additional activity beyond their function as CDK inhibitors. In this regard, Cip/Kip proteins might play a crucial role in the regulation of transcription factors and their expression aberrancies could result in tumor development 789.
Altogether, since P21 and P27 have crucial roles in cell cycle regulation, it is likely that leukemic cells profit from changes in their gene expression. Since few studies have been conducted to evaluate the importance of such proteins in ALL patients, we therefore investigated the expression patterns of pivotal cell cycle genes (P21 and P27) in newly diagnosed ALL cases.
Methods
Patients and healthy people
In the present study, we assessed 38 bone marrow (BM) and 14 peripheral blood (PB) samples from newly diagnosed ALL patients, and 13 BM and PB samples from healthy individuals. Patient consent was obtained, according to the institutional ethics guidelines for the study (IR.SBMU.RETHECH.REC.1396.800). Diagnosis of ALL was confirmed by morphology characteristics, immunophenotyping, and molecular analysis. For collection of control samples, we utilized BM and PB specimens from patients with no hematologic malignancies and whom were referred to the lab for routine checkup by physicians.
RNA extraction and cDNA synthesis
Total cellular RNA was extracted using RNeasy Kit (Qiagen, Germany). The integrity of RNA was measured by the NanoDrop (Thermo Scientific, Wilmington, North Carolina, USA). High purity (OD 260/280 nm ratio 1.8) was observed for all samples. Subsequently, cDNA was synthesized from 1 g of RNA to a final volume of 20 L by means of a cDNA Synthesis Kit (ThermoFisher Scientific, Waltham, MA, USA). An equal amount of cDNA from controls and patients (50 ng of cDNA) was used as a substrate for qRT- PCR amplification.
Real-Time PCR (qRT-PCR)
The primers used in the present study were designed via Oligo 7.56 software and the data were checked in the NCBI Blast database. The primer sequences were:
P27 forward (5- AGCAATGCGCAGGAATAAGGAAGC-3) and
P27 reverse (5- CACAGAACCGGCATTTGGGGAACC -3),
P21 forward (5- ACTAGGCGGTTGAATGAGAG -3) and
P21 reverse (5- GAGAGGAAAAGGAGAACACG-3), and
ABL forward (5-AGTCTCAGGATGCAGGTGCT-3) and
ABL reverse (5-TAGGCTGGGGCTTTTTGTAA),
according to previous studies; ABL was selected as a housekeeping gene in this study 10.
Consequently, P21, P27 and ABL mRNA expression in the patient and healthy volunteer samples were analyzed by qRT-PCR (Rotor-Gene 6000, Qiagen, Germany). The components of the qRT-PCR reaction for each target consisted of 1 L of template target cDNA, 1 L of forward and reverse primer, 7L of Real Plus 2x Master Mix Green-Low ROX (Ampliqon, Denmark), and 6 L water for a total reaction volume of 15 L. For each qRT-PCR reaction, a standard curve was produced, using five consecutive 1:10 dilutions of a positive sample (1, 0.1, 0.01, and 0.001). The thermal cycler program for each reaction included an initial hold at 95°C for 10 minutes, followed by 40 cycles of denaturation at 95°C for 10 seconds and annealing/extension at 60°C for 30 seconds. The relative quantification of mRNA expression for each sample (fold change = FQ) was calculated using the Livak method (2) 11.
Statistical Analysis
The SPSS statistics software (version 16.0) and Graph Pad Prism software (version 6.07) were used for data analysis. In SPSS, LOG was used for normalizing distribution of (2) for performing statistical analyses. The Shapiro-Wilk test was applied to determine the normal distribution of the data. One-way ANOVA or Kruskal-Wallis for multi-state variables and t-test or Mann-Whitney U test for two state variables were used according to the Shapiro-Wilk test. Pearson’s chi-square test was performed to analyze correlations. Two-tailed P<0.05 was considered to be significant.
Results
Specification of patient samples
In this study, the expression levels of P21 and P27 were analyzed in peripheral blood and bone marrow specimens collected from 52 patients with ALL. Samples were of different age, sex, and distinct malignant morphological characteristics
Specification of newly diagnosed ALL patients
| Study population | |
| Age (years) | 1-89 |
| MaleSexFemale | 30 (57.70%)22 (42.30%) |
| Peripheral bloodSample typeBone marrow | 14 (26.92%)38 (73.08%) |
| RangeBlast percent | 20-98 |
P21 and P27 expression in ALL patients and normal controls
Our results revealed that P21 and P27 expression were decreased in ALL patients in comparison to the normal control group (8.33- and 1.69-fold change, respectively) Figure 1.

Relative P21 and P27 mRNA expression. The P21 fold change obtained 8.33 (P<0.05) in ALL patients compared with normal control group. The P27 fold change obtained 1.69 in ALL patients compared with normal control group.
Differential expression of P21 and P27 according to ALL immunophenotype groups
The differential expression of P21 and P27 between the different immunophenotype groups in ALL patient samples were evaluated by ANOVA test. Statistical analysis of P21 expression showed a significant difference between T-ALL and early pre B-ALL (P=0.03). However, P27 expression was not significantly different among the immunophenotype groups (P= 0.35) Figure 2.

Differential expression of P21 and P27 according to ALL immunophenotype groups. The ANOVA sample was applied to evaluation of differential expression of P21 and P27 between the different immunophenotype groups in ALL patient samples. Statistical analysis of P21 expression revealed a significant difference between T-ALL and early pre B-ALL (P=0.03). P27 expression revealed no significant difference between different immunophenotype groups (P= 0.35).

P21 and P27 expression in B-ALL patients and control group. P21 and P27 expression in B-ALL and control group was evaluated using t-test. P21 expression demonstrated statistically significant difference between B-ALL and control group (P= 0.001). P27 expression demonstrated no significant difference between B-ALL and control group (p value 0.47).
P21 and P27 expression in B-ALL patients and control group
P21 and P27 expression in B-ALL and control group were evaluated using t-test. P21 expression in the B-ALL group was significantly different that of the control group (P=0.001). P27 expression was not significantly different between the B-ALL group and the control group (P=0.47) Figure 3.
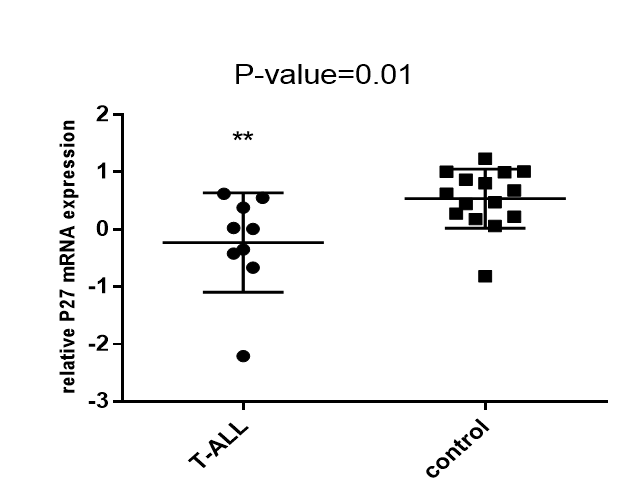
P27 expression in T-ALL patients and control group. P21 and P27 expression in T-ALL and control group was evaluated using t-test. Significant difference in P27 expression between T-ALL patients and control group was indicated (P=0.01).

Expression of P21 and P27 in T-ALL and B-ALL patients. The differential expression of P21 and P27 between T-ALL and B-ALL patient samples was evaluated by the t-test. Statistical analysis of P21 and P27 expression showed significant difference between T-ALL and B-ALL patients (P=0.03).
P21 and P27 expression in T-ALL patient group and control group
P21 and P27 expression in T-ALL group and control group were evaluated using t-test. There appeared to be a significant difference between the T-ALL patients and controls Figure 4.
Expression of P21 and P27 in T-ALL and B-ALL patients
The differential expression of P21 and P27 in the T-ALL patient samples versus B-ALL patient samples was evaluated by t-test. Statistical analysis of P21 and P27 expression showed a significant difference between T-ALL and B-ALL patients (P=0.03) Figure 5.
Discussion
Genetic abnormalities including gene mutations and trans-locations are insufficient to generate the whole leukemia characteristics in most patients. Indeed, both genetic and epigenetic modifications induce cooperating altered cell cycle progression and inhibition of apoptosis, which are required for establishment of neoplasms 121314. As an intricate process, cell cycle regulation can undergo different alterations resulting in cell proliferation abnormalities and cancer progression. Proper cell division is ensured by various mechanisms controlling cell cycle 1516. These mechanisms include CDK regulation through cyclins, CDK inhibitors, and phosphorylation 17. Studies have revealed that the expression of cell cycle regulators, such as P21 and P27, is relevant to aggressive tumor patterns in human malignancies 1819202122. Although cell cycle deregulation have been indicated in different types of cancers, there are insufficient studies evaluating the expression patterns of cell cycle regulators in ALL with variable outcomes. Therefore, we evaluated the expression of P21 and P27 as two main CDKIs involved in cell cycle regulation.
In the present study, we found that P21 and P27 expression levels were diminished by 8.33- and 1.69-fold, respectively, in ALL patient samples compared to the control group. Our results demonstrated a decreased expression of P21 in 94% and reduced expression of P27 in 50% of the ALL patients in our study.
In agreement with our results, Roman-Gomez (2002) has evaluated the P21 promoter methylation pattern in ALL patients; P21 hypermethylation was observed in 41% of their patients and was strongly correlated with decreased expression of P21 in cancerous cells 5. Pellikainen . (2003) demonstrated a decreased expression of P21 in their study on breast cancer samples (by IHC techniques), which was concordant with our results 23. Radosevic . (2001) also detected very low levels of P21 protein in their fresh AML samples 24. In agreement with our results, Zolota . detected P21 only in a small percentage of their BM samples. However, P27 expression was observed in a high percentage of their cases, which is in contrast with our findings 25. It seems that decreased expression of P21 as a tumor suppressor gene in leukemic cells is the result of activation of oncogenic pathways, and facilitates leukemic growth.
In a study by Letestu . (2004), P27 expression was evaluated in Mantle cell lymphoma (MCL) using western blot technique. The authors observed an absence of P27 expression in 72% of their patients while P27 median expression was observed in only 28% of patients. They also showed that survival of P27 negative patients was greater than that of P27 positive patients 26. Møller (1999) investigated P27Kip1 expression in non-Hodgkin’s lymphoma (NHL) patients by immunohistochemistry. They demonstrated that lower expression of P27Kip1 was predictive of poor survival in indolent and aggressive NHL 27. Chiarle . (2000) also investigated the expression of P21 and P27 in MCL. They observed loss of P27 in a high percentage of MCL cases due to proteasome-mediated degradation; the loss of P27 was associated with poor outcomes 28. Halvorsen (2003) also evaluated the expression of P27 and phosphatase and tensin homolog (PTEN) in primary prostatic carcinomas. In their study, P27 low expression was associated with high clinical stage of prostatic carcinoma 29.
Moreover, in an epigenetic analysis by Chim . (2005) and Takeuchi (2011) on cell cycle regulators, such as P21 and P27, in patients with acute leukemia, P21 hypermethylation was not detected and P27 methylation was rarely demonstrated 430. Winkler . (2010) also reported P27 overexpression in patients with CLL, which was associated with higher risk groups 31. Van de Putte . (2002) showed that P21 expression was increased in 20% of patients with early squamous cervical cancer, while P27 expression was decreased in 80% of their samples 32. In malignant ovarian tumors, Lee . (2011) showed P21 overexpression compared with borderline tumors. P27 expression was not significantly changed among benign, borderline, and malignant tumors in histologic evaluations of ovarian tumors. The authors showed that cytoplasmic P27Kip1 was relevant and associated with shorter overall survival in ovarian carcinomas 33.
The discrepancies among the reported studies possibly arise from the employment of different analysis methods, from protein degradation, and/or from unexpected functions. P21 and P27 are believed to exert their inhibitory functions through similar pathways as a result of high homology in their primary structures 34. It seems that P21 and P27 genes are subject to changes at the transcriptional level in ALL patients. Generally, P21 and P27 can be considered as strong tumor suppressors in ALL leukemic cells; their expression can, indeed, be influenced by epigenetic modifications or alternative mechanisms. It is postulated that P21 and P27 tightly regulate cell cycle such that their absence or aberration can result in proliferation and tumor growth in ALL. Moreover, the appropriate cellular localization of P21 and P27 can govern their tumor suppressor functions 353637.
In conclusion, limited studies are available concerning expression, regulation, and localization patterns of cell cycle regulators, such as P21 and P27, in hematologic malignancies. Future studies should be done to assess these genes not only at the transcription level but also at the post-transcriptional and translational levels.Evaluation of expression of these genes can enhance our knowledge about the molecular mechanisms involved in ALL formation and progression. Furthermore, these evaluations can help in developing ALL risk classifications precisely and in yielding more effective therapies for ALL patients.
Conclusions
Since P21 and P27 are able to influence the activity of both cyclins and CDKs, the decreased expression of these genes reduces P21- and P27- mediated suppressive effects on cyclins and CDKs. These events can, therefore, facilitate the activation of cyclins and CDKs, which could result in cancer progression in ALL patients.
Open Access
This article is distributed under the terms of the Creative Commons Attribution License (CCBY4.0) which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
List of abbreviations
ALL: Acute Lymphoblastic Leukemia; CDK: Cyclin-Dependent Kinases; CDKI: Cyclin-Dependent Kinase Inhibitor; CIP/KIP: CDK interacting protein/Kinase inhibitory protein ; CLL: chronic lymphocytic leukemia; INK4: Inhibitors of CDK4; MCL: Mantle cell lymphoma; NHL: non-Hodgkin’s lymphoma; QPCR: quantitative polymerase chain reaction
Ethics approval and consent to participate
Patient consent was obtained, according to the institutional ethics guidelines for the study (IR.SBMU.RETHECH.REC.1396.800)
Competing interests
The authors declare no conflict of interest.
Funding
Shahid Beheshti University of Medical Sciences, Shahid Beheshti, Iran.
Authors' contributions
All authors contributed to the design of the research. MKH, MHM, PKH and SL collected the
Data. HA, AGH and MA conducted analysis and interpretation of data. All authors drafted
The first version. MKH, MHM, PKH, SL, HA and ZKH edited the first draft. All authors reviewed, Commented and approved the final draft.
Acknowledgments
We would like to express our gratitude to the department of hematology and blood banking, Shahid Beheshti University of Medical Sciences and HSCT Research Center of Taleghani hospital.

