
Elevated expression of SKP2 correlates with poor prognosis in osteosarcoma: a bioinformatics analysis
- Department of Orthopedics, Zhongshan Hospital, Fudan University, Shanghai, China
- Department of Spinal Surgery, Institute of Trauma and Orthopedics, 108 Military Central Hospital, Hanoi, Viet Nam
- Department of Orthopedics, Shanghai Geriatrics Center, Fudan University, Shanghai, China
Abstract
Introduction: Osteosarcoma (OS) is the most prevalent kind of bone cancer, but the tumorigenesis and underlying molecular drivers of OS remain unknown. The instability of the cell cycle regulation system, which leads to uncontrolled cell proliferation, is a typical characteristic of carcinogenesis. This study aimed to investigate the expression of SKP2 in human OS and assessed its prognostic value in OS patients.
Methods: Three gene expression profile datasets GSE28424, GSE42352 and GSE21257 were obtained from the GEO database. The Wilcoxon rank-sum test was performed to evaluate the differential expression of the SKP2 gene between the OS and the control groups in the GSE28424 and GSE42352 datasets, and the correlation between SKP2 expression and Huvos grade was analysed in the GSE21257 dataset. Subsequently, PPI network, GO and KEGG analyses were constructed. Furthermore, survival analysis between high SKP2 expression group and low SKP2 expression group was performed using the Kaplan–Meier method and log-rank test. In addition, the UALCAN database was used to investigate the association between SKP2 expression and sarcoma.
Results: The expression level of SKP2 in OS cells was significantly higher than that in normal bones and mesenchymal stem cell samples. Furthermore, the level of SKP2 expression was observed to decrease as Huvos grade increased. The PPI network was established, and the top ten SKP2-related genes were identified, including CDKs (1, 2, 4, and 6), Cyclin A1-2, E1-2, D1, and CDKN1A. The survival analysis showed that the elevated SKP2 expression level was significantly related to the overall survival of OS patients.
Conclusion: Our work adds to our knowledge of SKP2's function in OS and suggests that it might be used as a therapeutic target in the future.
Introduction
Osteosarcoma (OS) is the most prevalent kind of bone cancer, mostly happening in young individuals aged 10 – 25 years and comprising over one-third of all primary bone malignancies1, 2, 3. The treatment for OS patients has progressed remarkably during the last four decades, and the advances in therapy, especially the introduction of chemotherapy as well as neoadjuvant chemotherapy followed by primary tumour resection, have led to a dramatically amelioration in the long-term prognosis of individuals without distal metastasis3, 4, 5. However, the prognosis for advanced patients remains poor, and early diagnosis has been restricted due to our limited understanding of the aetiology. Besides, the tumorigenesis and underlying molecular drivers of OS remain unknown. Thus, identifying novel effective biomarkers and drug targets is the key to understanding the development and progression of OS and to improve patient survival rates.
The instability of the cell cycle regulation system, which leads to uncontrolled cell proliferation, is a typical characteristic of carcinogenesis, and a variety of regulatory factors in the cell cycle are degraded through the ubiquitin-proteasome pathway. SKP2 (S-Phase Kinase-Associated Protein 2), also named FBXL1 (F-Box/LRR-Repeat Protein 1) or p45 (CDK2/cyclin A-associated protein p45), encodes for a 45 kDa protein that belongs to the F-box family, it could ubiquitinate and degrade numerous tumour suppressor proteins such as p27, and so plays a crucial role in regulating the cell cycle. SKP2 was initially identified as a pro-tumour factor in 1995, when it was discovered to be a crucial part for the S-phase of the cyclin A — CDK2 kinase in several transformed cells6. It was found that SKP2 was highly expressed and played a major role in carcinogenesis in numerous kinds of malignancies, including soft tissue sarcomas (STS)7, breast cancer8, hepatocellular carcinoma (HCC)9, pancreatic ductal adenocarcinoma (PDAC)10, astrocytic gliomas11. To our knowledge, no bioinformatics study has been conducted to investigate the involvement of SKP2 in OS. Hence, in the present study, we aimed to investigate the expression of SKP2 in human OS cell lines and samples and assessed its prognostic value in OS patients.
Methods
Microarray data
The Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) was used in this study. GEO, which is established and managed by the National Center for Biotechnology Information, National Library of Medicine, is an invaluable international resource that preserves and freely accesses comprehensive sets of microarray data submitted by researchers globally12, 13. The database includes data from a wide variety of technologies, such as Serial Analysis of Gene Expression, DNA microarray, protein array, high-throughput nucleic acid sequencing, and Real-Time PCR14, 15.
The search strategy we employed to identify OS-related datasets in the GEO database was as follows: “(osteosarcoma OR OS) AND Expression profiling by array[DataSet Type] AND Homo sapiens[Organism]”. Datasets GSE2842416, GSE4235217 and GSE2125718 were eventually included and extracted after a thorough search.
The platform for GSE28424 is GPL13376 — Illumina HumanWG-6 v2.0 expression beadchip, which contains nineteen OS cell lines and four normal bones. The platform for GSE42352 is GPL10295 — Illumina human-6 v2.0 expression beadchip, which comprises nineteen OS samples and twelve mesenchymal stem cell (MSC) samples used as controls. The GSE21257 dataset, which is also based on the GPL10295 platform, has fifty-three OS samples with complete clinicopathological characteristics.
Data processing
The R software (version 4.0.5) was used to acquire and process the expression profiling data. First, the raw data of all datasets was obtained using the GEOquery package (v2.58.0) and normalized and standardized using the limma package (v3.46.0). Then, the Wilcoxon rank-sum test was performed to evaluate the differential expression of SKP2 gene between the OS and the control groups in the GSE28424 and GSE42352 datasets. In the GSE21257 dataset, the samples were divided into a high expression group and low expression group based on the median value of SKP2 expression, and survival analysis was performed using the Kaplan–Meier method and log-rank test.
UALCAN database
UALCAN (http://ualcan.path.uab.edu) is a web-portal with user-friendly interactive features for conducting in-depth analysis of gene expression data from The Cancer Genome Atlas (TCGA)19. The mRNA expression of SKP2 in sarcoma tissues and adjacent normal tissues were explored. In addition, we also investigated the effect of SKP2 expression level on sarcoma patient survival through the UALCAN database by comparing the overall survival between the SKP2 high expression group and the SKP2 low/medium expression group.
Protein-Protein Interaction (PPI) network construction
The STRING database, which is a tool for retrieving and displaying the genes that a query gene appears within clusters on the genome20, was employed to establish the PPI network of SKP2 protein and other proteins. The PPI network was then visualized using the Cytoscape program21. Furthermore, Cytohubba, a Cytoscape plug-in, was performed to detect the hub genes that are significantly related to SKP2.
Functional and pathway enrichment analysis
Gene Ontology (GO) analysis aims to standardize the representation of gene and gene product properties across species, comprising cell components (CC), biological process (BP) and molecular function (MF)22. Kyoto Encyclopaedia of Genes and Genomes (KEGG) analysis serves as a combined database resource for biologically interpreting totally sequenced genomes, the process to map genes in the genome to generated pathway maps23.
GO and KEGG were utilized to probe the molecular mechanisms and pathways related to SKP2 for the development and prognosis of OS, and the visualization of the enrichment analyses was conducted through the cluster-Profiler package (v3.18.1), the GOplot package (v1.0.2), and the enrichplot package (v1.10.2) in the R language software.
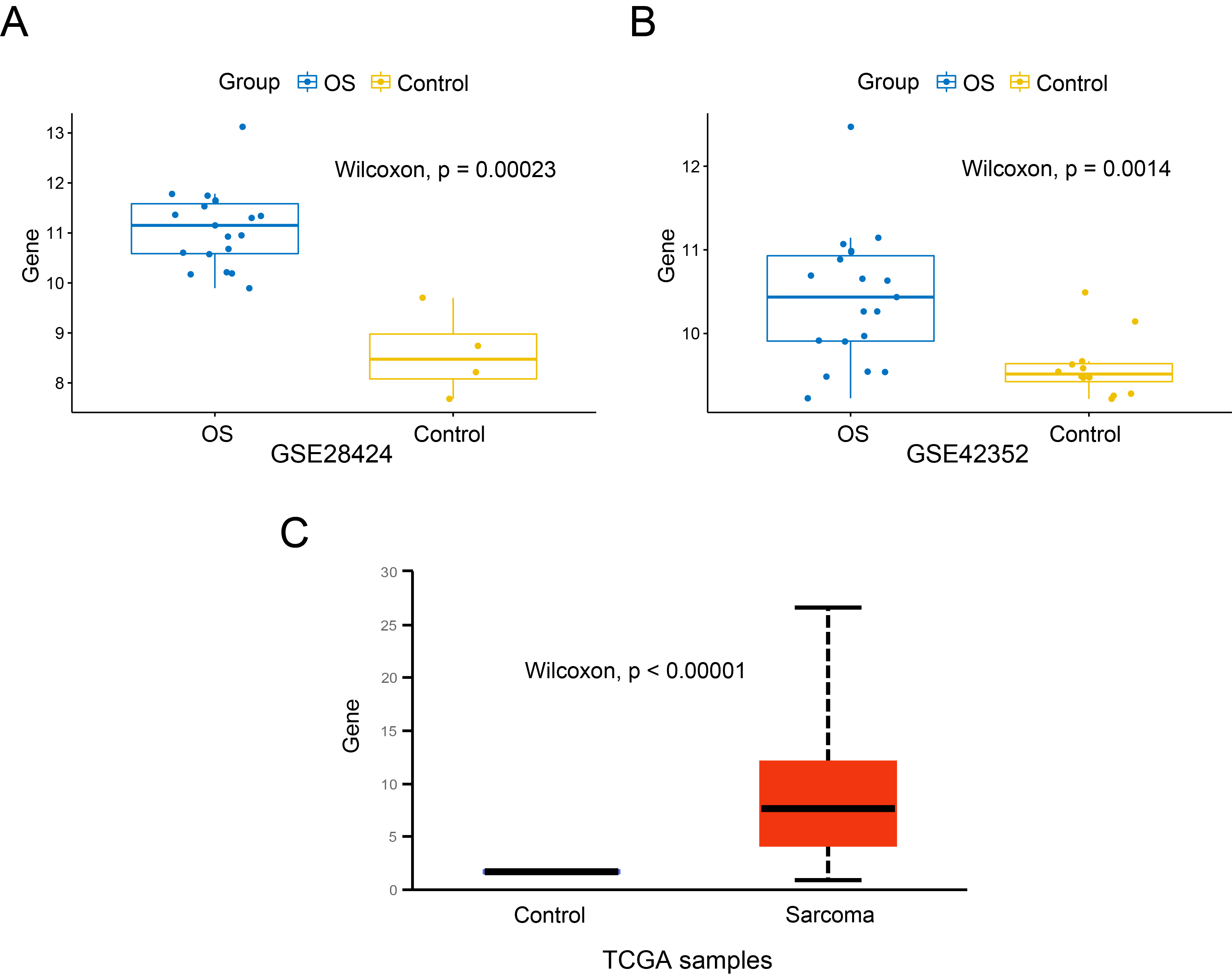
Expression of SKP2 mRNA in OS and control cells. A. GSE28424 dataset, OS cell lines and normal bone cells; B. GSE42352 dataset, OS samples and MSC samples; C. TCGA database, sarcoma tissues and the corresponding normal tissues.
Results
Increased SKP2 expression in OS
The expression of SKP2 in OS cells and control cells was compared. In GSE28424 (Figure 1A), the expression level of SKP2 in OS cell lines was significantly higher than that in normal bones (P < 0.001). We also found that SKP2 expressed highly in OS samples compared to MSC samples in GSE42352 (P = 0.0014), as shown in Figure 1B. Besides, the expression difference of SKP2 between the tumour tissues of sarcoma and the corresponding normal tissues in the TCGA project was additionally evaluated through the UALCAN web portal (Figure 1C), and it was statistically significant (P < 0.001). These findings indicate that SKP2 is expressed at an obviously high level in OS cells.

SKP2 expression in osteosarcoma with different Huvos pathological grades in the GSE21257 dataset.
Relationship between SKP2 expression and Huvos grade
The Huvos grading system is widely used in the chemotherapy assessment of OS, and it is an important parameter for predicting long-term prognosis. Grades I and II are considered to be poor in response to chemotherapy, while grades III and IV are regarded to have a favourable response. We used the GSE21257 dataset to perform a correlation analysis between SKP2 expression and Huvos grade. The level of SKP2 expression was observed to decrease as Huvos grade increased (P < 0.05) (Figure 2), indicating that the expression level of SKP2 falls as the rate of tumour necrosis tissue increases the following chemotherapy. This suggests that SKP2 might be used to predict the effectiveness of OS chemotherapy.
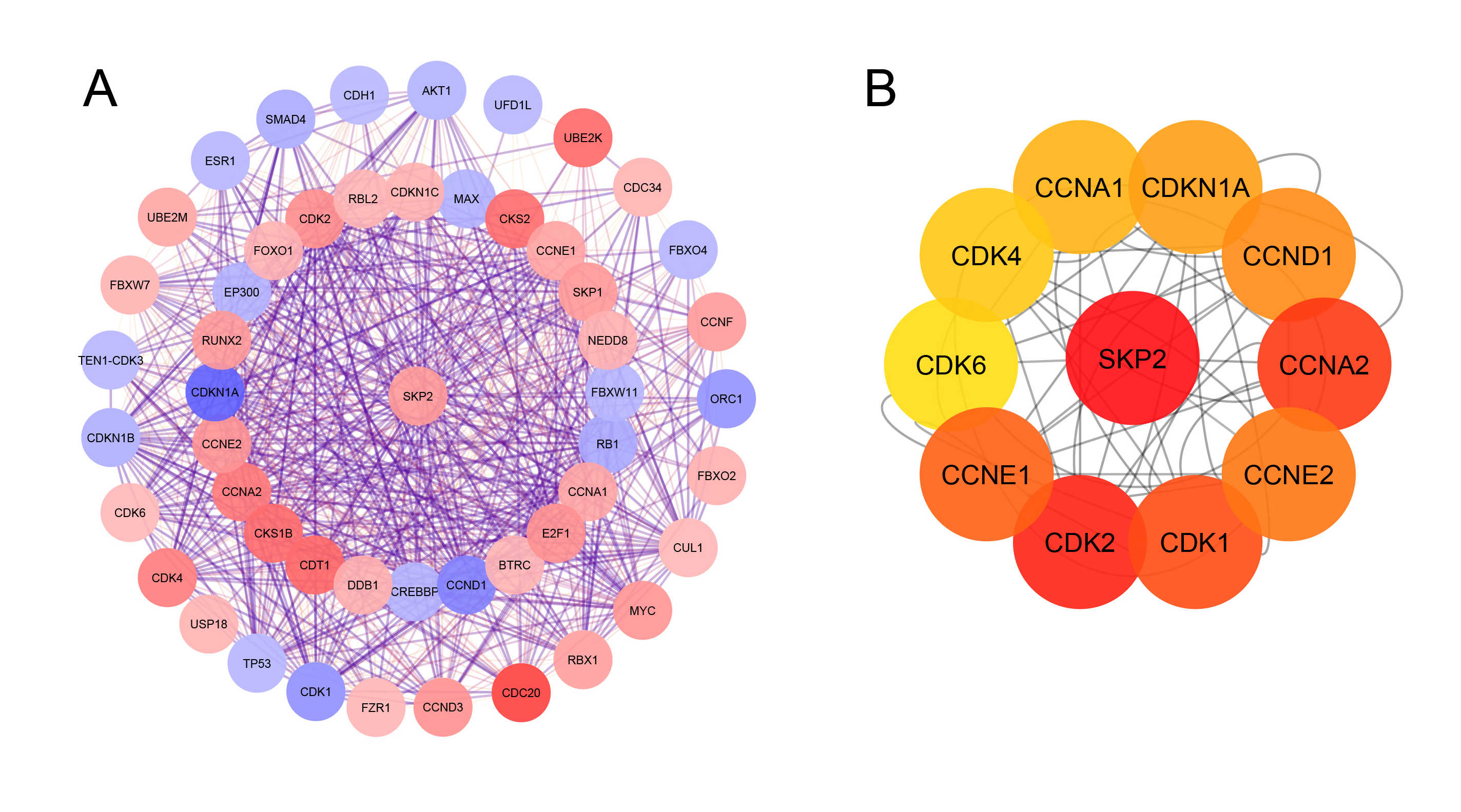
Protein-ProteinInteraction (PPI) network. A. PPI network of SKP2 and the other related proteins; B. the top ten SKP2-related genes using cyto Hubba plug-in.
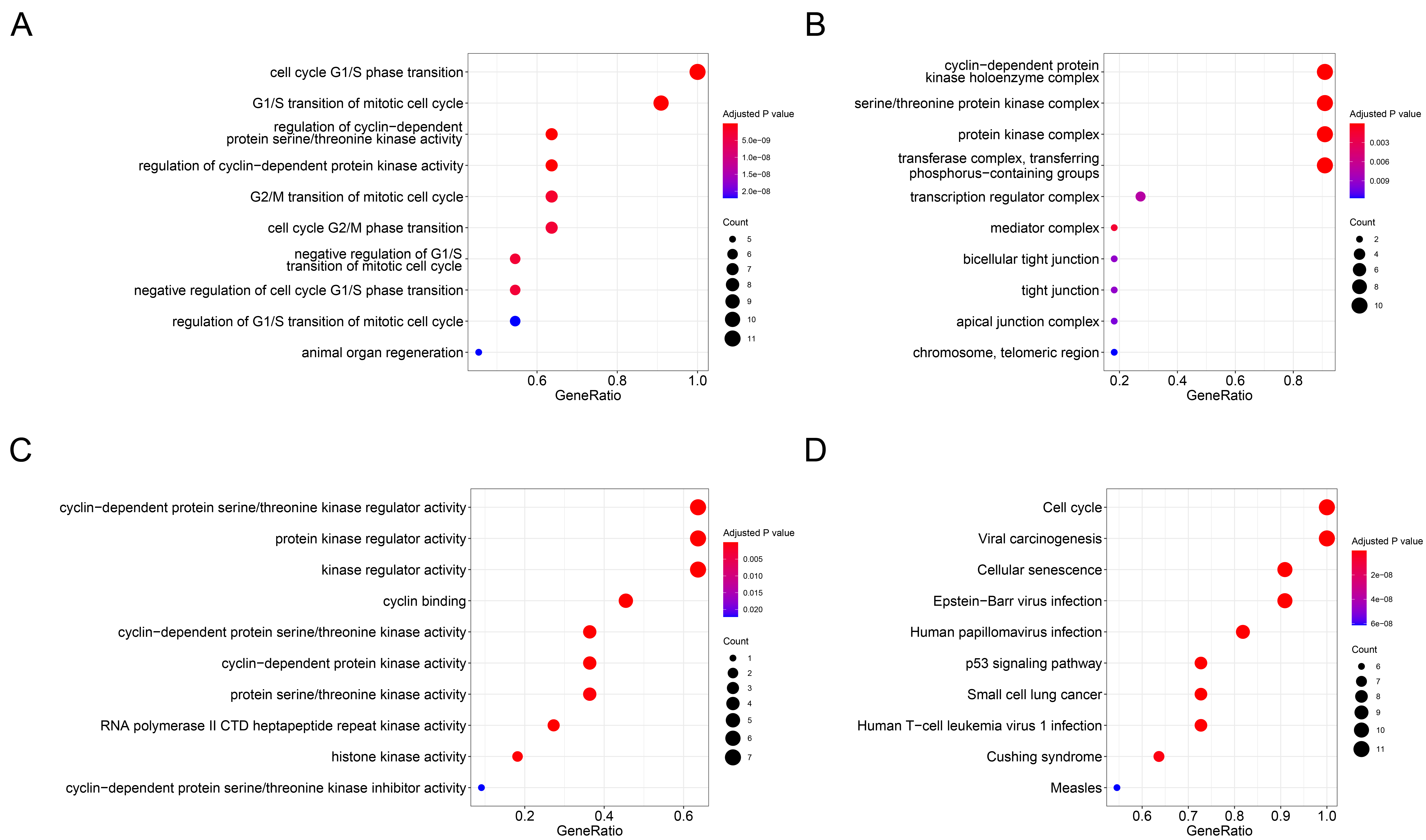
Functional enrichment analysis. A. Biological processes (BP) analysis; B. Cell composition (CC) analysis; C. Molecular function (MF)analysis; D. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis.
Top 5 results of Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses
|
GO/KEGG |
ID |
Description |
BgRatio |
P value |
Adjusted P value |
q value |
Count |
|---|---|---|---|---|---|---|---|
|
BP |
GO:0044843 |
cell cycle G1/S phase transition |
310/18866 |
1.98E-20 |
1.11E-17 |
5.05E-18 |
11 |
|
BP |
GO:0000082 |
G1/S transition of mitotic cell cycle |
287/18866 |
6.16E-18 |
1.73E-15 |
7.88E-16 |
10 |
|
BP |
GO:0000079 |
regulation of cyclin-dependent protein serine/threonine kinase activity |
102/18866 |
3.55E-14 |
6.56E-12 |
2.99E-12 |
7 |
|
BP |
GO:1904029 |
regulation of cyclin-dependent protein kinase activity |
106/18866 |
4.68E-14 |
6.56E-12 |
2.99E-12 |
7 |
|
BP |
GO:0000086 |
G2/M transition of mitotic cell cycle |
254/18866 |
2.33E-11 |
2.61E-09 |
1.19E-09 |
7 |
|
CC |
GO:0000307 |
cyclin-dependent protein kinase holoenzyme complex |
43/19559 |
9.35E-27 |
2.99E-25 |
1.08E-25 |
10 |
|
CC |
GO:1902554 |
serine/threonine protein kinase complex |
89/19559 |
2.47E-23 |
3.96E-22 |
1.43E-22 |
10 |
|
CC |
GO:1902911 |
protein kinase complex |
104/19559 |
1.27E-22 |
1.35E-21 |
4.90E-22 |
10 |
|
CC |
GO:0061695 |
transferase complex, transferring phosphorus-containing groups |
253/19559 |
1.19E-18 |
9.55E-18 |
3.46E-18 |
10 |
|
CC |
GO:0016592 |
mediator complex |
41/19559 |
0.000233 |
0.001491 |
0.00054 |
2 |
|
MF |
GO:0016538 |
cyclin-dependent protein serine/threonine kinase regulator activity |
50/18352 |
2.35E-16 |
7.29E-15 |
3.22E-15 |
7 |
|
MF |
GO:0019887 |
protein kinase regulator activity |
185/18352 |
3.01E-12 |
3.90E-11 |
1.72E-11 |
7 |
|
MF |
GO:0030332 |
cyclin binding |
30/18352 |
3.77E-12 |
3.90E-11 |
1.72E-11 |
5 |
|
MF |
GO:0019207 |
kinase regulator activity |
216/18352 |
9.00E-12 |
6.98E-11 |
3.08E-11 |
7 |
|
MF |
GO:0004693 |
cyclin-dependent protein serine/threonine kinase activity |
29/18352 |
1.65E-09 |
8.51E-09 |
3.75E-09 |
4 |
|
KEGG |
hsa04110 |
Cell cycle |
126/8095 |
8.35E-21 |
5.18E-19 |
1.76E-19 |
11 |
|
KEGG |
hsa05203 |
Viral carcinogenesis |
204/8095 |
1.99E-18 |
6.18E-17 |
2.10E-17 |
11 |
|
KEGG |
hsa04218 |
Cellular senescence |
156/8095 |
5.73E-17 |
1.18E-15 |
4.02E-16 |
10 |
|
KEGG |
hsa05169 |
Epstein-Barr virus infection |
202/8095 |
8.08E-16 |
1.25E-14 |
4.25E-15 |
10 |
|
KEGG |
hsa04115 |
p53 signaling pathway |
73/8095 |
4.76E-15 |
5.91E-14 |
2.01E-14 |
8 |

Kaplan–Meier analysis for overall survival based on SKP2 expression. A. Between high expression group and low expression group in OS. B. Between high expression group and low/medium expression group insarcoma.
PPI network construction
The PPI network created by STRING, which consisted of 51 nodes and 709 edges, was visualized using Cytoscape to explore the functional connections between SKP2 and the other proteins related to it (Figure 3A). Subsequently, as shown in Figure 3B, the top ten related genes identified using the Maximal Clique Centrality method in the Cytohubba plug-in were CDKs (1, 2, 4, and 6), Cyclin A1-2, E1-2, D1, and CDKN1A.
GO and KEGG enrichment analyses
Enrichment analyses of GO and KEGG pathway for SKP2 and its top ten associated genes were performed and displayed using the R software packages clusterProfiler, GOplot, and enrichplot (Figure 4,
Relationship between SKP2 expression and prognosis in OS patients
We further studied the role of SKP2 in the prognosis of OS patients. In the GSE21257 dataset, based on the cut-off value, patients were split into two groups: low expression group (26 patients) and high expression group (27 patients). The Kaplan-Meier curve and log-rank test shown that the elevated SKP2 expression level was significantly related to the overall survival (P < 0.01) of OS patients (Figure 5A). Moreover, the dataset in UALCAN also indicated that there was a significant difference in survival time between the SKP2 high expression group (65 patients) and the SKP2 medium/low expression group (194 patients) (P = 0.017), as shown in Figure 5B. Taken together, OS patients with a high level of SKP2 expression were predicted to have a poor prognosis.
Discussion
OS is one of the most prevalent bone cancers, affecting primarily adolescents and children and has a 5-year overall survival rate of approximately 50% — 70%1. The histology of OS is very variable, and its genetic structure is unstable, resulting in poor survival for the patients24. Hence, new prognostic biomarkers are required to assist patients with OS to improve their outcomes.
There have been several bioinformatics studies on OS in the past. HLA-DRA, EGR1, CXCL10, MYC, and CXCR4 were discovered to be hub normal-primary differentially expressed genes (DEGs) in OS25, 26. Besides, Zhang .27measured the amounts of tumour-infiltrating immune cells in the OS tumour microenvironment via the ESTIMATE algorithm-based immune score, and they detected 191 immune-related DEGs, with PPARG, IGHG3, PDK, CD209, and CCL8 ranking first through fifth. Another research studied the differential expression of circRNA in OS, they determined that circ_20403 and circ_2137 were highly upregulated and circ_24831 and circ_32279 were highly downregulated in OS samples, these circRNAs were significantly associated with BP and MF28.
SKP2 encodes for a type of Leucine-rich repeat protein that belongs to the F-box family29. SKP2 was initially recognized as an important cell cycle regulator because the ubiquitin-mediated degradation of several regulators of cell cycle required the promotion of SKP230, 31. Owing to the fact that the majority of its ubiquitinated and degraded substrates are tumour suppressors, SKP2 is classified as an oncoprotein. Besides, many studies also suggested that SKP2 functions as an oncogene in human malignancies.
In a large-scale analysis, elevated SKP2 expression in patients with STS was demonstrated as an independent risk factor for disease-specific survival in females and those who did not undergo chemo- or radiotherapy32. Another research showed that medium-to-high SKP2 expression in the nucleus in ≥ 10% of the cells was recognized in 37% of the total cases of STS and was more frequent in the high-grade group than in low-grade group7. Regarding breast cancer, cytoplasmic over-expression of SKP2 is presented in invasive ductal carcinoma, positively correlated with tumour grade and tumour size and inversely correlated with patient outcomes33. In HCC, Lee . revealed that LKB1 polyubiquitination mediated by Skp2 was crucial for tumour development in vivo34; in addition, enhanced SKP2 expression was shown to be negatively associated with apoptotic process and prognosis and positively associated with the increased level of tumour cells and microvascularization9. About PDAC, SKP2 tended to overexpress in patients with higher tumour grade, more lymph node metastases, greater amount of lymphatic permeation, and worse survival10.
In this study, the role of SKP2 in OS was investigated using a bioinformatics approach. According to our findings, all three datasets GSE28424, GSE42352 and TCGA determined that SKP2 was upregulated in OS, this result was consistent with prior research. When compared to normal human osteoblasts or human MSC-derived osteoblasts, Zhang .35 found that the levels of Skp2 mRNA were significantly higher in both conventional and patient-derived OS cell lines. In OS cells, elevated expression of SKP2 was determined to stimulate cell proliferation and cell cycle progression, reduce apoptosis, and accelerate cell invasion and migration36. Notably, Nagao .37 revealed that overexpression of SKP2 induced overexpression of GLI2 in OS cells, resulting in enhancing cell proliferation and promoting cell cycle progression, showing that Skp2 was an important link in the control of cell growth in OS cells. Another study discovered that SKP2 interacts with its substrate p27 to promote OS development and stemness38. Furthermore, SKP2 might be used as a potential prognostic and pro-metastatic biomarker. The findings of our survival study determined that SKP2 was tightly related to prognosis in OS patients, with greater levels of SKP2 expression having a worse prognosis. Similar to this result, in a study of metastasis-free survival analysis in 88 untreated high-grade OS patients, the high Skp2 group had a poorer prognosis than the low Skp2 group35. In addition, Liu .39 also demonstrated that OS patients with lung metastasis with high SKP2 TPM (Transcripts Per Million) had a worse survival than those with low SKP2 TPM.
Moreover, on the basis of established criteria, the top ten SKP2-related genes were selected as hub genes for further investigation, which were CDK1-2, 4, and 6, Cyclin A1-2, D1, E1-2, and CDKN1A. These genes are all closely related to the cell cycle, which is a critical process in malignant transformation. Disturbances in the cell cycle are frequently documented in tumorigenesis40. A vast body of literature has shown CDK (cyclin-dependent kinase) family members exert as oncoproteins and are deregulated in various malignancies, changes in CDK activity are frequently involved in tumour-associated cell cycle abnormalities41. CDK6 overexpression may promote OS cell proliferation and migration42, whereas SKP2 overexpression could increase the level of CDK6 through its substrate p27. Regarding CDK2, suppressing or downregulating it in OS cell lines could stop them from proliferating43, 44. Cyclin family regulates the cell progression through triggering CDK enzymes or synthesis-related enzymes for cell cycle45. Various prior reports have demonstrated that many members of the cyclin family, including cyclin A, D, and E, are significantly expressed and/or involved in cell proliferation in OS46, 47, 48, 49, 50, 51. Li . discovered that hsa_circ_0003732 may enhance OS cell proliferation by increasing cyclin A2 expression through miR-54552. Besides, numerous biomarkers, such as URG4, JARID1B, lncRNA LINC01296, promote OS cell proliferation, metastasis and cell cycle via cyclin D147, 48, 53. Additionally, cyclin E1 overexpression was identified as associated with clinicopathology and a potential marker predicting outcome for OS patients50.
The Huvos system was used to observe the effect of chemotherapy on high-grade OS in the GSE21257 dataset and in the present study. The Huvos grading system is a histologic evaluation system for bone sarcomas in which the level of necrosis in relation to the percentage of remaining viable tumour is determined for histologic assessment purposes using a semiquantitative method54. Grades 1 – 4 indicate necrosis of < 50%, 50% — 90%, 90% — 99%, and 100%, respectively, and higher grades are regarded to have a favourable response to chemotherapy. Our finding showed that individuals with high grades had a significantly low expression of SKP2 than those with low grades, suggesting that chemotherapy may exert its effect through inhibition of SKP2 expression. Given the crucial participation in regulating cancer progression, we believed that SKP2 might be a new target for the therapeutic treatment of OS. A prior report showed that downregulating the expression of SKP2 by PPARG could effectively induce impaired cell proliferation and apoptosis, suggesting that targeting SKP2 expression with PPARG agonists could be a viable treatment approach55. Hirotsu .56 discovered that suppressing SMO with cyclopamine promoted OS cell cycle arrest as well as prevented cell development by decreasing the transcription of Skp2. Moreover, upregulation of miR-506 induces Skp2 to be reduced, which stimulates apoptosis and slows motility in OS cells57. Interestingly, both SKP2 inhibitors (compound C1, pevonedistat and FKA) and SKP1-SKP2 pocket inhibitor (compound C25) have anti-proliferative effects on pRB/p53 double-deficient OS cells38. Conclusively, developing SKP2 inhibitors is a feasible and promising method for treating OS.
Conclusions
In conclusion, the expression and prognostic value of SKP2 in OS were formulated in the present study. SKP2 was overexpressed in OS cell lines and samples and correlated with poor outcomes in OS patients. Given its importance in the cell cycle, our research suggests that SKP2 might be an important biomarker in OS tumorigenesis as well as a potential target for drug development and treatment.
Abbreviations
BP: Biological process, CC: Cell components, CDK: Cyclin-dependent kinase, DEG: Differentially expressed gene, GEO: Gene Expression Omnibus, GO: Gene Ontology, HCC: Hepatocellular carcinoma, KEGG: Kyoto Encyclopedia of Genes and Genomes, MF: Molecular function, MSC: Mesenchymal stem cell, OS: Osteosarcoma, PDAC: Pancreatic ductal adenocarcinoma, PPI: Protein-Protein Interaction, SKP2: S-Phase Kinase-Associated Protein 2, STS: Soft tissue sarcomas, TCGA: The Cancer Genome Atlas, TPM: Transcripts Per Million
Acknowledgments
None.
Author’s contributions
Juan Li and Hong Lin, conception and design; Hong Lin, administrative support; Tien-Manh Hoang, Minh-Tien Nguyen and Weisin Chen, collection and assembly of data; Tien-Manh Hoang, Minh-Tien Nguyen, Chenyang Zhuang, Hanquan Wang, data analysis and interpretation. All authors wrote the article. All authors read and approved the final manuscript.
Funding
This work was supported by the Non-profit Central research Institute Fund of Chinese academy of Medical sciences [2020-JKCS-013]; and Three-Year Initiative Plan for Strengthening Public Health System Construction in Shanghai [GWV-10.2-XD11].
Availability of data and materials
All data were obtained from the GEO database (http://www.ncbi.nlm.nih.gov/geo/). The authors will also provide researchers with the full dataset upon request.
Ethics approval and consent to participate
The study was exempted from institutional review board approval since the clinical data was obtained from a publicly accessible database.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.

