Rapid screening B-cell Acute Lymphoblastic Leukemia withBCR::ABL1 fusion transcript by aberrant immunophenotype
- Department of Hematology, Cho Ray Hospital, Ho Chi Minh City 700000, Viet Nam
- The Laboratory D Unit, Department of Biochemistry, Cho Ray Hospital, Ho Chi Minh City 700000, Viet Nam
- Department of Ultrasonography and Functional Test, Cho Ray Hospital, Ho Chi Minh City 700000, Viet Nam
Abstract
Introduction: B-cell acute lymphoblastic leukemia (B-ALL) with Philadelphia (Ph) chromosome or BCR::ABL1 fusion transcript is a high-risk subtype in hematological malignancy. It is crucial to identify this cytogenetic abnormality because it can help clinicians decide the best treatment regimen quickly and prolong overall survival for patients. The gold standard for detecting the BCR::ABL1 fusion transcript and Ph chromosome is real-time quantitative polymerase chain reaction (RQ-PCR) and fluorescence in situ hybridization (FISH) techniques, respectively. In this study, we aimed to screen B-ALL with BCR::ABL1 fusion transcript by aberrant immunophenotype during the routine diagnosis process.
Methods: We divided forty B-ALL patients into two groups based on the presence of the BCR::ABL1 fusion transcript and Ph chromosome. Immunophenotype analysis and white blood cell (WBC) count were performed for all patients. Comparing the expression level of each antigen between the two groups, we recorded the aberrant immunophenotype related to BCR::ABL1 rearrangement. Bayesian Model Averaging (BMA) was used to find an optimal model to predict the BCR::ABL1 fusion transcript in B-ALL disease.
Results: Besides WBC count (p = 0.004), the presence of three aberrant immunophenotype markers—CD66c (p < 0.001), CD25 (p = 0.002), and CD38 (p = 0.003)—was related to BCR::ABL1 rearrangement. BMA analysis proposed a model to predict B-ALL with BCR::ABL1 fusion transcript using three variables: WBC count and the expression intensity of CD38 and CD66c. The predictive model using WBC count, CD38, and CD66c showed an Area Under the Curve (AUC) value of 0.938 (CI95%: 0.868–1.000), a sensitivity of 85% (CI95%: 70%–100%), and a specificity of 90% (CI95%: 75%–100%) (p < 0.001).
Conclusion: The combined model using WBC count, CD38, and CD66c proposed in this study was useful in predicting B-ALL with BCR::ABL1 fusion transcript.
Introduction
B-cell acute lymphoblastic leukemia (B-ALL) is a hematological malignancy characterized by the development and proliferation of lymphoblasts committed to the B-cell lineage, arrested at an early stage of B-cell maturation1. According to the World Health Organization (WHO) classification of tumors of hematopoietic and lymphoid tissues in 2016, B-ALL with recurrent cytogenetic abnormalities is a major concern for clinicians, specifically t(9;22) translocation creating the Ph-chromosome or BCR::ABL1 fusion transcript, as this represents a high-risk subtype2. In addition, Ph-chromosome or BCR::ABL1 fusion transcript is the most common cytogenetic abnormality, accounting for approximately 20% to 30% of adult ALL cases overall3, 4. Before the tyrosine kinase inhibitor (TKI) era, treatment for adult B-ALL with Ph-chromosome was dismal because the disease-free survival (DFS) and overall survival (OS) were short5, 6, 7. Since TKI was approved, chemotherapy combined with TKIs became the first-line therapy for the disease, achieving complete response (CR) and improving DFS and OS for patients8, 9, 10. Therefore, determining the presence of BCR::ABL1 fusion transcript when diagnosing B-ALL is crucial11.
Some research has shown B-ALL disease characterized by aberrant immunophenotypes that can be detected using flow cytometry. These aberrant immunophenotypes may correlate with specific cytogenetic abnormalities. Tabernero et al. recorded that CD10, CD13, CD34, and CD38 are related to the BCR::ABL1 gene rearrangements in B-ALL disorder12. Owaidah . reported that the expression levels of CD25 and CD66c were predictive of the presence of BCR::ABL1 rearrangement in B-ALL disease13. Tang . concluded that the BCR::ABL1 fusion transcript and CD66c exhibit high concordance in minimal residual disease detection of adult B-ALL14. No research in our country has mentioned aberrant immunophenotypes suggesting the presence of BCR::ABL1 rearrangement in B-ALL disorder until now, so we decided to conduct this study and identify the associated factors related to this cytogenetic abnormality.
Methods
Patients
Forty patients aged sixteen or older were chosen for this study. They were newly diagnosed with B-cell acute lymphoblastic leukemia (B-ALL) according to the WHO classification of tumors of hematopoietic and lymphoid tissues in 2016. Among the 40 patients, 20 tested positive for both Ph-chromosome and BCR::ABL1 fusion transcript, while the remaining 20 tested negative for both. All patients underwent comprehensive diagnostic evaluations, including complete blood count, morphological assessment, immunophenotyping, cytogenetic analysis, and molecular biology testing. All results were fully documented. The cytogenetic and molecular biology techniques followed standard protocols. The patients were enrolled at Cho Ray Hospital from March 2021 to September 2022. The study was approved by the Cho Ray Hospital Institutional Review Board (approval number 1170-BVCR-HĐĐĐ), and each patient provided signed informed consent.
Immunophenotype
Analysis We used the ClearLLab 10C system (Beckman Coulter, California, USA) for the diagnosis and classification of immunophenotype in patients, including ClearLLab 10C B cell tube (Cat no B96805) and M2 cell tube (Cat no B96808) according to the manufacturer's recommendations. Additionally, a single staining tube including CD66c (Cat no IM2039U), CD25 (Cat no IM0479U), CD22 (Cat no A60791), CD45 (Cat no B36294) was used, along with another tube including cyTdT (Cat no IM3524U), cyMPO (Cat no IM3455U), cyCD79a (Cat no A60793), cyCD3 (Cat no A66329), CD45 (Cat no B36294), to complement the diagnostic panel. Samples were acquired on the Navios EX cytometers using acquisition protocols provided by the manufacturer. The results of the immunophenotype analysis are collected using Kaluza software version 2.0 and analyzed by a professional hematologist.
Molecular and Cytogenetic Analysis
The t(9;22) translocation or Ph-chromosome was detected in the bone marrow sample using the FISH technique with the Vysis BCR/ABL1/ASS1 Tri-color DF FISH Probe Kit (Cat no 05N5420, Abbott Molecular, Illinois, USA) according to the manufacturer's instructions. Briefly, the white blood cells were collected and incubated with a 0.075 M potassium chloride (KCl) solution and Carnoy's fixation solution. After that, the cells were dropped on the marked area of a charged slide and incubated with a 10 µl probe mixture at 75°C/3 mins and 37°C/16–20 hours. Finally, the slide was washed with SSC/NP-40 solution and stained with DAPI-II solution before being analyzed using the Bioview fluorescence microscope system (Abbott Molecular, Illinois, USA). Slides were automatically scanned on the system, and the results were analyzed by professional molecular biology and genetics technicians. Positive results are calculated as the percentage of abnormal cells over the total number of analyzed cells (at least 200 cells) based on signal images captured by the system.For the BCR::ABL1 fusion transcript detection, total RNA from the bone marrow sample was extracted using the QIAamp RNA Blood Mini Kit (Cat no 52304, Qiagen, Hilden, Germany). The BCR::ABL1 fusion transcript was confirmed by the RQ-PCR technique using the ipsogen BCR::ABL1 Mbcr IS MMR DX Kit (Cat no 670823, Qiagen, Hilden, Germany) for the e13a2 and e14a2 transcript (major variant) and the ipsogen BCR::ABL1 mbcr (Cat no 670023, Qiagen, Hilden, Germany) for the e1a2 transcript (minor variant) according to the manufacturer's instructions. PCR reactions were performed on the RotorGene Q 5Plex HRM platform and analyzed using RotorGene Q 2.0 software.
Statistical Analysis
The Chi-square or Fisher’s exact tests were used to compare frequencies, while the Kruskal–Wallis rank test was used to compare the density expression of each antigen between groups. The Bayesian Model Averaging (BMA) statistic was applied to identify the markers associated with BCR::ABL1 rearrangement and the optimal diagnostic model. Logistic regression was employed to construct the receiver operating characteristic (ROC) curve and define the cut-off point, sensitivity, specificity, and the value under the ROC curve (area under the curve: AUC) of each antigen and the optimal diagnostic model for B-ALL with BCR::ABL1 fusion transcript. All data analyses were completed using R statistical software v.3.5.1 (R Foundation, Austria). A p < 0.05 was considered statistically significant.

Comparative box plot of WBC count between groups B-ALL with BCR::ABL1 (right) and non-BCR::ABL1 (left) fusion transcript. The boxplot shows the difference in WBC count between two groups has statistically significant (p = 0.004).
Abbreviations: -ALL: B-cell Acute Lymphoblastic Leukemia; BCR::ABL1: Breakpoint Cluster Region - Abelson 1; WBC: White Blood Cell
Results
Complete blood count analysis in study groups
When analyzing the complete blood count testing, leukocytosis was recorded in both study groups (Figure 1). In the BCR::ABL1 negative group, leukocytosis increased slightly with a median WBC count of 18.7 × 10/L. In contrast, the median of WBC count in the BCR::ABL1 positive group recorded 113.9 × 10/L, which was higher than in the BCR::ABL1 negative group (p = 0.004). So, the difference in WBC count between the two study groups suggested the ability to use WBC count as a predictive factor of BCR::ABL1 fusion transcript in B-ALL.

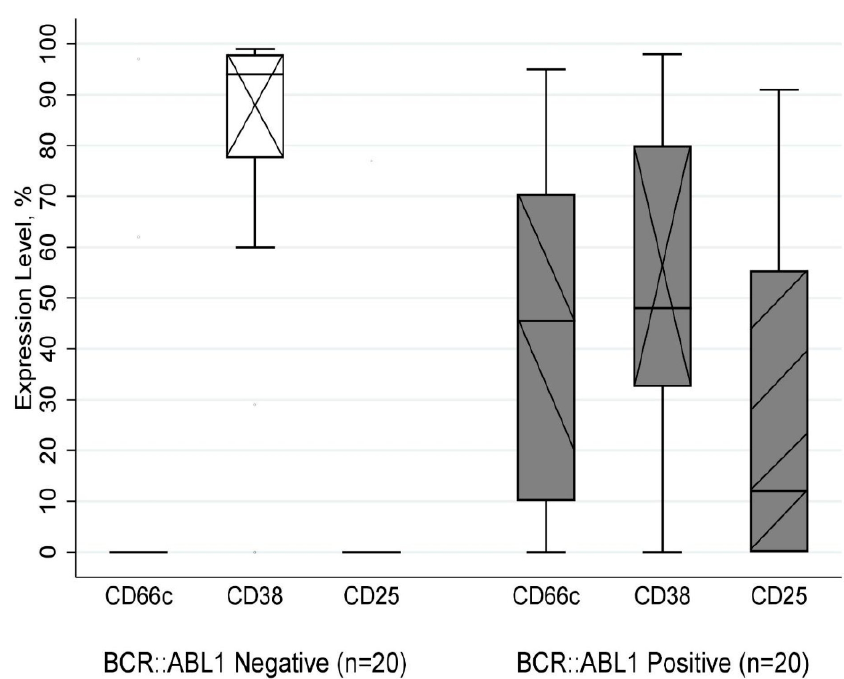
Expression level of CD66c, CD38, CD25 between groups B-ALL with BCR::ABL1 (right), and non-BCR::ABL1 (left) fusion transcript. The boxplot shows the difference in expression level of CD66c (p < 0.001), CD38 (p = 0.003), and CD25 (p = 0.002) between two groups has statistically significant. Abbreviations: B-ALL: B-cell Acute Lymphoblastic Leukemia; BCR::ABL1: Breakpoint Cluster Region - Abelson 1
Antigen expression between groups
All patients in the two groups had expressed CD19 combined with at least one other B-lineage marker (CD20, CD22, and cyCD79a), and all were negative for cyCD3 and cyMPO (Data not shown). We also recorded that B-ALL with BCR::ABL1 rearrangement share similar immunophenotype of CD10 (p = 0.384), CD19 (p = 0.877), CD20 (p = 0.341), CD22 (p = 0.704), CD34 (p = 0.396), cyCD79a (p = 0.713), CD123 (p = 0.892), CD200 (p = 0.506), and HLA-DR (p = 0.588) with B-ALL non-BCR::ABL1 fusion transcript. Interestingly, as shown in Figure 2, we found that the expression level of CD25 (p = 0.002) and CD66c (p < 0.001) in BCR::ABL1 positive patients were higher than in BCR::ABL1 negative patients when analyzing the expression of cell antigen. In contrast, the CD38 (p = 0.003) expression level in the positive group is lower than in the other.
Factors selection and combined model in rapid screening BCR::ABL1 fusion transcript in B-ALL
In this study, we diagnosed B-ALL by complete blood count, morphology, and flow cytometry panel using 24 immunophenotype markers. The presence of BCR::ABL1 fusion transcript was detected by RQ-PCR testing. By analyzing many factors related to BCR::ABL1 fusion transcript in B-ALL, we found that four potential factors: WBC count (p = 0.004), CD25 (p = 0.002), CD38 (p = 0.003), and CD66c (p < 0.001), which were closely associated with BCR::ABL1 rearrangement in B-ALL.
In the univariable logistic regression, B-ALL with BCR::ABL1 fusion transcript was differentiated from B-ALL non-BCR::ABL1 with an accuracy of 76.5% (AUC = 0.765, cut-off: ≥ 38.6 × 10/L) by WBC count, 72.2% by CD25 (AUC = 0.722, cut-off: ≥ 24% cells), 77.8% by CD38 (AUC = 0.778, cut-off: ≤ 84% cells), and 80.8% (AUC = 0.808; cut-off: ≥ 20% cells) by CD66c (
Diagnostic values of each factor for B-ALL with BCR::ABL1 fusion transcript
|
Factor |
Cut-off |
AUC (CI 95%) |
Sensitivity, % (CI 95%) |
Specificity, % (CI 95%) |
|
WBC |
≥ 38.6 x 109/L |
0.765 (0.611-0.919) |
75.0 (55.0-90.0) |
80.0 (60.0-95.0) |
|
CD25 |
≥ 24% cells |
0.722 (0.599-0.846) |
50.0 (30.0-70.0) |
95.0 (85.0-100.0) |
|
CD38 |
≤ 84% cells |
0.778 (0.623-0.932) |
70.0 (50.0-90.0) |
80.0 (60.0-95.0) |
|
CD66c |
≥ 20% cells |
0.808 (0.674-0.941) |
75.0 (55.0-90.0) |
90.0 (75.0-100.0) |

The diagnostic power of WBC count, CD38, and CD66c in combination. BMA analysis shows the Area Under the Curve (AUC) value of the Combine Model (0.938) is much higher than three factors including WBC count (0.765), CD38 (0.778), and CD66c (0.808) when diagnosing B-ALL with BCR::ABL1 rearrangement.
Abbreviations: BMA: Bayesian Model Averaging; WBC: White Blood Cell; AUC: Area Under the Curve; B-ALL: B-cell Acute Lymphoblastic Leukemia; BCR::ABL1: Breakpoint Cluster Region - Abelson 1
The BMA analysis suggested a combined model of three factors WBC count, CD38, and CD66c. The probability of the appearance of these factors being related to BCR::ABL1 fusion transcript in B-ALL were 91.6%, 72.2%, and 100% respectively. This model was selected for rapid screening BCR::ABL1 fusion transcript in B-ALL because it had the lowest Bayesian Information Criterion (BIC) score (-108.5) and the highest posterior probability (50.2%). Due to the low probability of occurrence, the BMA statistics did not include CD25 (43.8%) in the combined model. It both simplified the model and increased the accuracy of the model (Data not shown).
The multivariable analysis showed that the combined model of three factors WBC count, CD38, and CD66c resulted in a significantly increased accuracy value (AUC=0.938, 95% CI, 0.868–1.000, p < 0.001) (Figure 3). The sensitivity and specificity of this model in diagnostic B-ALL with BCR::ABL1 fusion transcript were up to 85% (CI 95%: 70.0–100.0) and 90% (CI 95%: 75.0–100.0), respectively.
Discussion
Flow cytometry is a diagnostic tool that is not only necessary in the classification of hematological malignancy but also in monitoring Minimal Residual Disease (MRD)15, 16. The most valuable advantage of this method is that it provides results quickly, making it ideal for the rapid screening of B-ALL with the BCR::ABL1 fusion transcript. In our country, there are few medical centers capable of performing both Flow cytometry and Molecular Biology - Genetic testing, especially for diagnosing B-ALL with BCR::ABL1 rearrangement. Therefore, the suggestion of BCR::ABL1 fusion transcript in B-ALL through aberrant immunophenotype markers in these cases is helpful for clinical physicians.
We assessed the expression level of aberrant immunophenotype markers and showed that CD66c and CD25 expression were highly related to the BCR::ABL1 fusion transcript in B-ALL disease. Paietta et al. previously reported that the expression of CD25 appeared to become a surrogate marker for the presence of BCR::ABL1 fusion transcript17. Besides, Sugita . reported CD66c predominantly expressed on the surface of Ph-chromosome-positive ALL cells18. However, we recorded an intermediate expression level of CD38 in our study, which was correlated to the BCR-ABL1 rearrangement in B-ALL disease. It was in contrast with the expression level of CD25 and CD66c. Hrusak and Porwit-MacDonald mentioned the intermediate expression of CD38 in B-ALL with t(9;22) or BCR::ABL1 rearrangement19. In addition, we found that the median WBC count is 113.9 x 10⁹/L in B-ALL with the BCR::ABL1 positive group. Previous research has shown that WBC count is moderately elevated in B-ALL with BCR::ABL1 disease, with reported values of 31.5 x 10⁹/L in China20, 26.9 x 10⁹/L in Japan21, and 35.8 x 10⁹/L in Korea22. The WBC count in our study is significantly higher than those reported in Japan, Korea, and China, indicating that our patients often presented with a high disease burden at diagnosis, a characteristic feature of B-ALL with BCR::ABL1 rearrangement used in risk stratification. It suggested that WBC count is a valuable factor when diagnosing B-ALL with BCR::ABL1 fusion transcript, not only for detecting the abnormal cytogenetic but also as a prognostic factor, as shown by previous studies23, 24.
Many studies have identified the aberrant immunophenotype in B-ALL disorder25, 26, 27, 28, but there is a lack of studies that combine multiple factors and rapidly suggest the presence of the BCR::ABL1 fusion transcript in B-ALL. Using BMA statistics, we have proposed an optimal model to predict the presence of the BCR::ABL1 fusion transcript in B-ALL based on the expression of CD38, CD66c, and WBC count at diagnosis. Since flow cytometry can provide quick results, we suggested using this model as the initial step in diagnosing B-ALL with the BCR::ABL1 rearrangement. Additionally, a multicolor flow cytometer device and an optimized panel of CDs can serve as a tool for rapid screening of B-ALL with the BCR::ABL1 fusion transcript. Subsequently, confirmation of the BCR::ABL1 fusion transcript using FISH and RQ-PCR techniques must be completed, following the current guidelines recommendation23, 24.
Conclusions
In conclusion, the results of this study demonstrated that WBC count and the expression levels of CD25, CD38, and CD66c in flow cytometry analysis are related to the presence of the BCR::ABL1 fusion transcript in B-ALL disease. Moreover, an optimal model combining WBC count, CD38, and CD66c has shown promising potential in using flow cytometry to predict the BCR::ABL1 fusion transcript in B-ALL disorder.
In this study, we present evidence of a highly comparable profile of cell antigens between B-acute lymphoblastic leukemia (B-ALL) cases exhibiting the BCR::ABL1 fusion transcript and those without it. We underscore the importance of utilizing alternative cut-off points rather than a fixed threshold of 20% events for more effective classification in practical applications. However, the study is limited by a small sample size and was conducted as a single-center retrospective analysis. Future research should focus on large-scale investigations to confirm the effectiveness of these findings.
Abbreviations
AUC: Area under the curve, B-ALL: B-cell Acute Lymphoblastic Leukemia, BCR::ABL1: Breakpoint Cluster Region - Abelson 1, BMA: Bayesian Model Averaging, CD: Cluster of differentiation, cy: Cytoplasmic, FISH: Fluorescent in situ hybridization, HLA-DR: Human Leukocyte Antigen DR, MPO: Myeloperoxidase, Ph-chromosome: Philadelphia chromosome, RNA: Ribonucleic acid, ROC: Receiver operating characteristic, RQ-PCR: Real-time quantitative polymerase chain reaction, t(9;22): Translocation (9;22), TdT: Terminal deoxynucleotidyl transferase, TKI: Tyrosine kinase inhibitor, WBC: White blood cells.
Acknowledgments
The authors thank all patients for their cooperation, generosity, and patience. The authors thank all the doctors, nurses, and medical technologists in the Department of Hematology for their professional assistance. In particular, the authors are grateful to Professor Vu Anh Hoang from the Center for Molecular Biomedicine at the University of Medicine and Pharmacy for his enthusiastic support in completing the manuscript.
Author’s contributions
TTT and HPM are senior authors who conceived and designed the study; TMT, HTV, LVP, and ThTT selected patients for the study and collected clinical data; HTHV, NTBC, and TMT carried out the immunophenotype analysis; HCP, HTLN, PHL, and HTV carried out the molecular biology and cytogenetic analysis; HPM, TTP processed and performed the data analysis. HPM wrote the manuscript. All authors read and approved the final manuscript.
Funding
None.
Availability of data and materials
Data and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The study was approved by Cho Ray Hospital Institutional Review Board (approval number 1170-BVCR-HĐĐĐ) and signed informed consent from each patient.
Consent for publication
Not applicable.
Declaration of generative AI and AI-assisted technologies in the writing process
No generative AI or AI-assisted technologies were used in the writing, editing, or creation of this manuscript. All work was performed by the authors.
Competing interests
The authors declare that they have no competing interests.

