Unveiling COVID-19: Exploring Associated Comorbidities and Diagnostic Innovation
- RNA Biology Lab, CSIR Institute of Genomics and Integrative Biology, Delhi-110025, India
- Molecular Biology and Genetics Unit, Jawaharlal Nehru Centre for Advanced Scientific Research, Bengaluru, Karnataka-560064, India
- Department of Microbiology, University of Delhi, South Campus, Delhi-110021, India
- Department of Microbiology, Shaheed Rajguru College of Applied Sciences for Women, University of Delhi, Vasundhara Enclave, Delhi-110096, India
- Trivitron Healthcare Pvt Ltd, AMTZ Campus, Pragati Maidan, Visakhapatnam-530031, India
- Department of Botany, University of Delhi, Delhi, India
- Department of Botany, Swami Shraddhanand College, University of Delhi, India
- Lady Irwin College, University of Delhi, India
Abstract
The outbreak of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has led to a rapid global spread, causing significant morbidity, mortality, and an economic downturn. The virus exhibits a unique genome organization comprising various proteins crucial for genome replication and host-cell receptor interaction, thereby affecting organs such as the lungs, kidneys, and heart. Prompt detection and the development of therapeutic strategies are crucial due to the virus’s severity and associated comorbidities. Well-established methods such as RT-PCR and ELISA, along with cutting-edge approaches like salivary diagnostics, nucleic acid sequence-based amplification (NASBA), and CRISPR technology, are essential for both diagnosis and a deeper understanding of the viral life cycle. Additionally, vaccination campaigns have proven pivotal in reducing infection rates and fatalities worldwide. Although definitive treatments remain elusive, vaccination remains a successful strategy for preventing illness and mitigating severe outcomes. This review aims to explore these techniques in depth, emphasizing their roles in COVID-19 detection, systemic post-infection effects, and potential pharmacological targets in future drug development. By thoroughly understanding the epidemiology, pathogenicity, symptomatology, diagnosis, and prophylaxis of CoVs, we can more effectively counteract the virus’s impact on global healthcare.
Introduction
Coronavirus disease 2019 (COVID-19) has triggered a serious global outbreak. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel mutated variant of the existing coronaviruses (CoVs) on Earth1, 2. Globally, the virus has rapidly spread throughout the world, infecting 775,552,205 people and causing 7,050,201 fatalities as of June 8, 2024, bringing the world to a halt (Figure 1 )3.
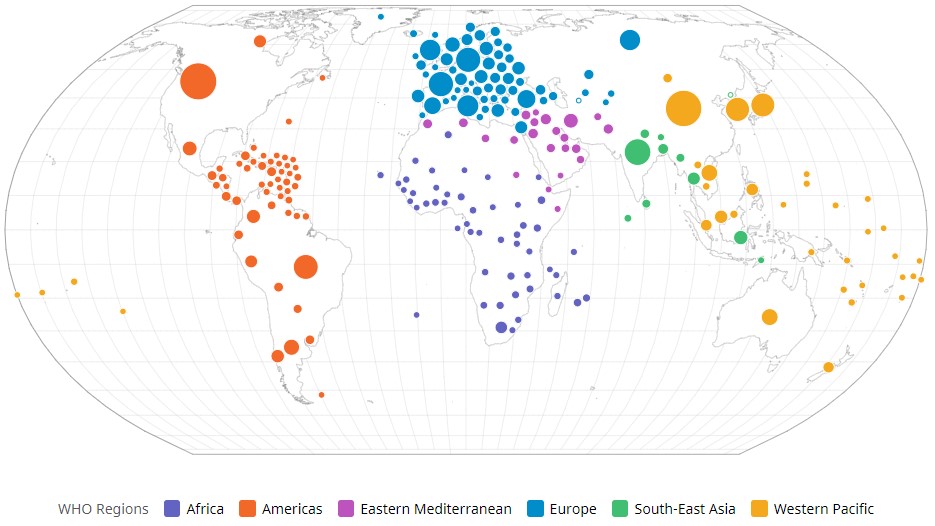
The global prevalence of COVID-19 cases. WHO COVID-19 Dashboard. Geneva: World Health Organization, 2020. Available online: https://covid19.who.int/ Accessed on 8 June 2024
CoVs are known to have an enveloped, single-stranded, positive-sense RNA genome. CoVs, including Middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV, and SARS-CoV-2, carry signature markers of previous zoonotic events. The current pandemic strain is the ninth documented coronavirus to infect humans4, 5, 6. Currently, MERS-CoV is producing sporadic outbreaks in the Middle East, and SARS-CoV caused an epidemic in China in 2003.
SARS-CoV-2 infection can range from lethal to asymptomatic. Common symptoms include cough, fever, muscle pain, fatigue, dizziness, and loss of taste and smell7. However, infection of the lower respiratory tract by any of these three CoVs can lead to acute respiratory distress, multiple organ failure, and sepsis, which can be fatal8. Individuals with diabetes, obesity, advanced age, cardiovascular disease, or certain cancers are more susceptible to severe coronavirus infection9.
Since the onset of the COVID-19 pandemic, especially in the post-pandemic era, there has been a dramatic surge in diagnostic innovations geared toward low-resource settings. The unmet demand during the pandemic has acted as a catalyst for accessible, affordable, and rapid diagnostic tools aimed at broader public use10. Point-of-care (POC) tests have been integrated with microfluidics and biosensors, utilizing artificial intelligence for real-time analysis, especially in remote settings. Various global health organizations, including the WHO, continue to update targeted product profiles and essential diagnostic lists, prioritizing equitable access and diagnostic innovations for vulnerable communities, particularly given COVID-19's disproportionate impact on these settings11.
Although a vast body of literature on COVID-19 exists, further exploration of this disease is still needed. Moreover, despite the novelty of the infection and many global clinical trials for potential therapeutics, including vaccines, there is still no absolute cure. However, to some extent, the use of certain emergency-use vaccines, along with social distancing measures, has helped mitigate disease spread.
SARS CoV-2: Origin and its evolutionary journey
In November 2002, the first major CoV outbreak was reported in the Foshan region of China, known as SARS-CoV. Subsequently, the MERS-CoV outbreak was documented in Saudi Arabia in June 2012.
The seventh human coronavirus (hCoV) in the last 20 years, SARS-CoV-2 (20A.EU2), was identified in 2019 during the pneumonia outbreak in Wuhan, Hubei Province, China.
SARS-CoV-2 is a highly contagious and divergent strain that originated from SARS-CoV, infecting human hosts and continuing to evolve10. However, various coronaviruses have also been documented, including alpha-CoV (229E, NL63) and beta-CoV (OC43, HKU1)3.
SARS-CoV-2 belongs to the beta-CoV group and seems to have originated from a bat coronavirus, as indicated by a 96% RATG13 sequence homology12, 13 (Figure 2 ). Bioinformatics studies also reveal that the SARS-CoV-2 genome shares approximately 79.5% sequence homology with the previously identified SARS-CoV and a 99% sequence identity with pangolin-derived beta-CoVs, suggesting pangolins as possible intermediate hosts in the transmission cycle13. Hence, the SARS-CoV-2 pandemic highlights the importance of zoonotic reservoirs for lethal CoVs, which continue to unveil hidden secrets over time and underline the need for clear boundaries between humans and wildlife12.
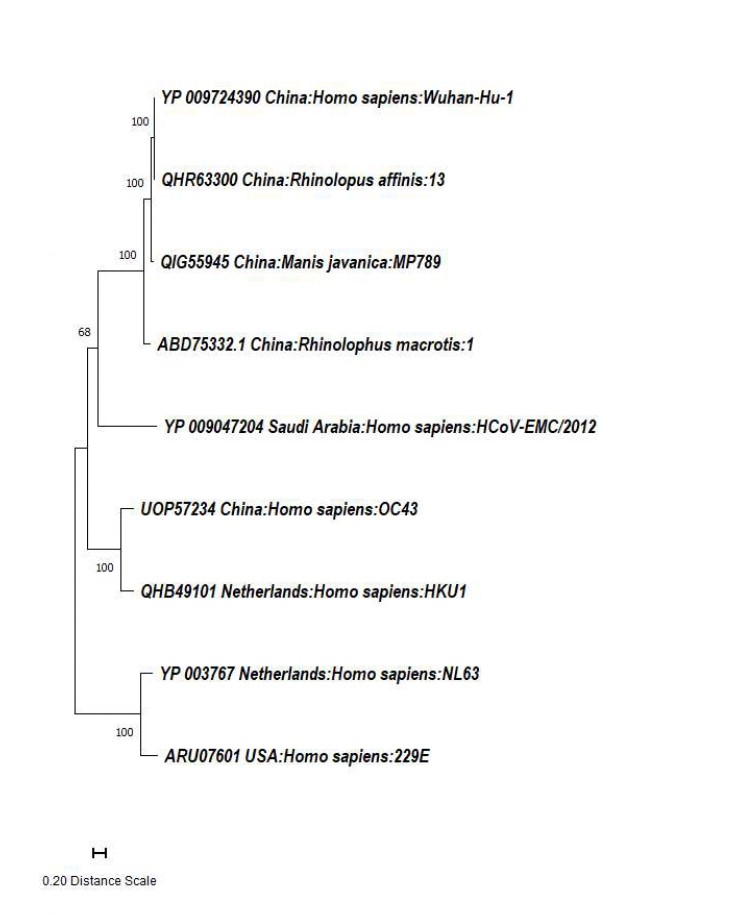
Phylogenetic analysis of SARS CoV-2 spike proteins.
A phylogenetic tree was constructed to illustrate the relationships among spike protein sequences of various CoV strains, including SARS-CoV-2, obtained from the National Center for Biotechnology Information (NCBI). A Maximum Likelihood (ML) and JTT matrix-based phylogenetic tree was generated using Molecular Evolutionary Genetics Analysis (MEGA 11). The resulting topology had the highest log likelihood value (-20118.86). Branch lengths were scaled and expressed as the number of amino acid substitutions at each site. Nine amino acid sequences were examined in this study, resulting in a final dataset of 1494 positions.
The most recent JN.1 variant strain of SARS-CoV-2, descended from the BA.2.86 Omicron lineage, was first identified in the United States in September 2023. India reported its first case of this variant in Kerala on December 8, 2023. The variant was designated a "variant of interest" by the World Health Organization (WHO) due to its higher transmission rates than previous strains14, 15, 16, although it posed a low estimated risk to global public health.
Overview of SARS-CoV-2
Ecological Framework
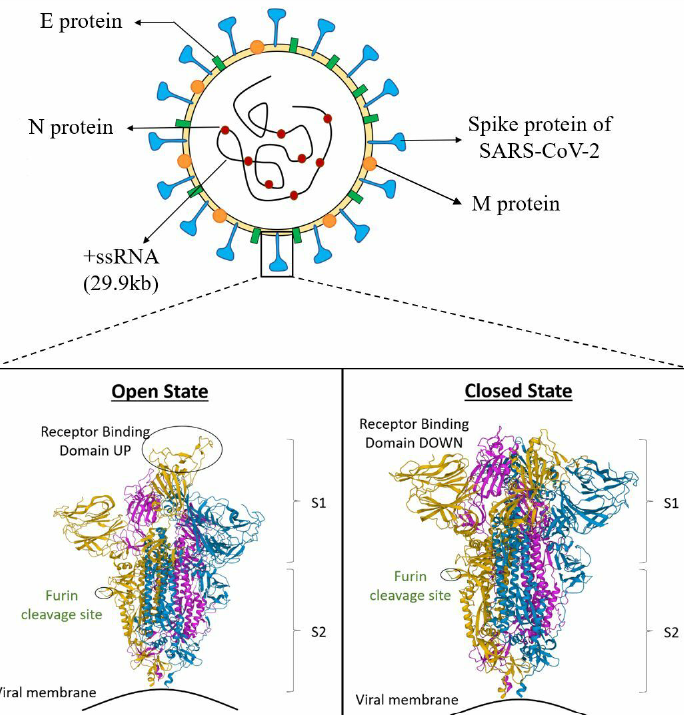
The virus is named for its distinctive spike proteins that protrude from its envelope, creating a crown-like appearance. SARS-CoV-2 possesses a helical capsid containing single-stranded RNA as its genetic material. The capsid is formed via phosphorylation of the nucleocapsid (N) protein. The structural proteins in the envelope include spike (S) glycoprotein, hemagglutinin esterase (HE), membrane (M) protein, and envelope (E) protein (Figure 3 )17.
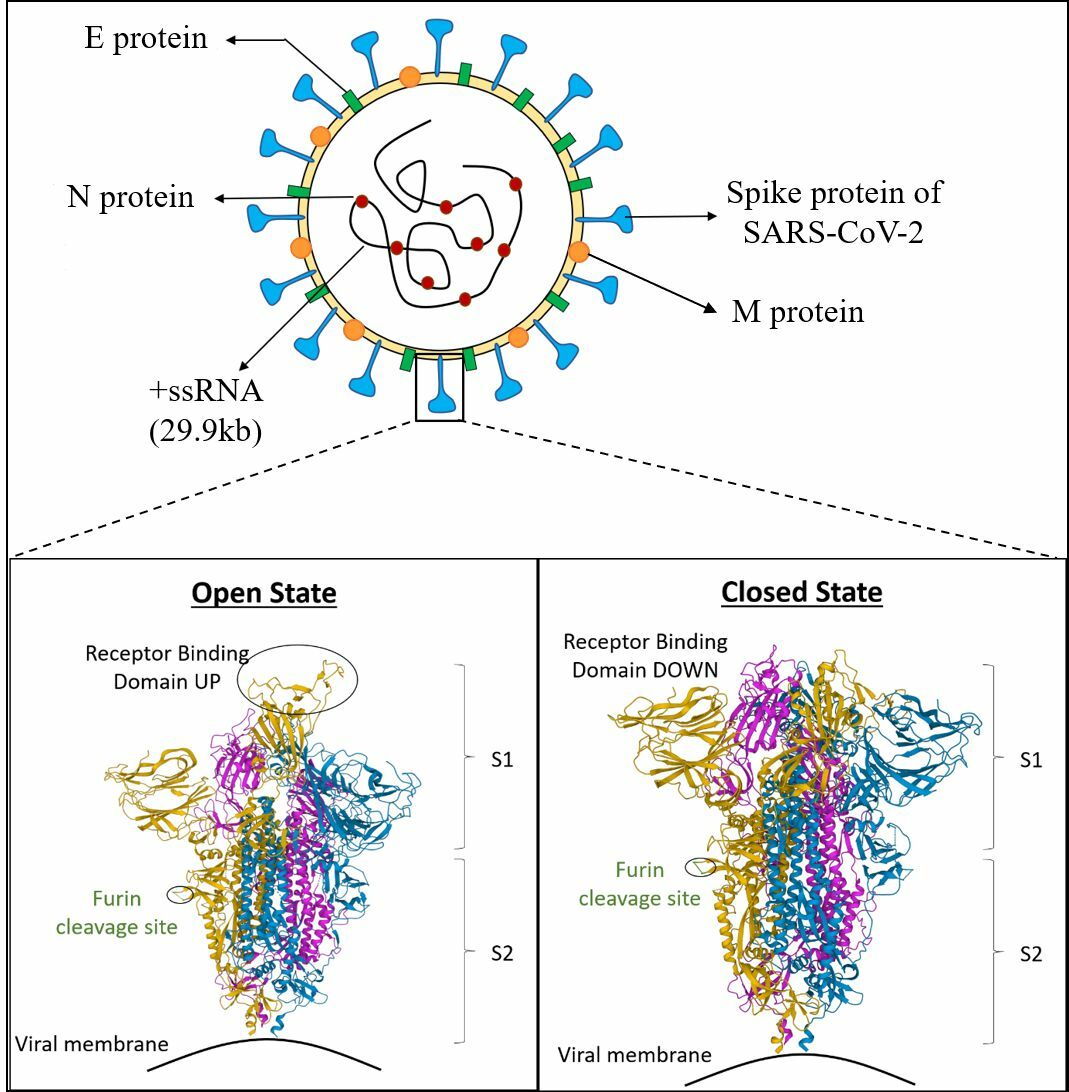
Schematic representation of SARS CoV-2 representing different proteins and 3D view of Receptor Binding Domain (RBD); Cartoon structure of closed (PDB ID, 6ZGI) and (PDB ID, 6ZGG) S protein of SARS CoV-2. The green colored portion represents the furin cleavage site. The section highlighted in yellow, blue, and pink color depict A, B and C chains, respectively. The Protein Data Bank (PDB) structures are taken from the Research Collaboratory for Structural Bioinformatics-PDB (RCSB-PDB) site. PDB. DOI (6ZGI): 10.2210/pdb6ZGI/pdb; PDB DOI (6ZGG): 10.2210/pdb6ZGG/pdb.
Infection Cycle
In typical coronaviruses (CoVs), the S protein of SARS-CoV-2 facilitates host cell receptor recognition and initiates the viral infection cycle. The S2 subunit comprises the fusion peptide, heptapeptide repeat sequences (HR1 and HR2), a transmembrane domain, and cytoplasmic domains, arranged systematically. These components are crucial for viral fusion and entry into the host cell, making them an attractive drug target13, 18. The fusion peptide is an oligopeptide composed of water-repelling glycine or alanine residues that disrupt the lipid bilayer of the host cell’s plasma membrane, aiding in membrane fusion. Meanwhile, HR1 and HR2 facilitate viral fusion and entry by forming the six-helical bundle structure. They are part of a repetitive HPPHCPC heptapeptide sequence, where H, P, and C stand for hydrophobic, polar (hydrophilic), and charged residues, respectively19. As the receptor-binding domain (RBD) binds to angiotensin-converting enzyme 2 (ACE2), the S2 subunit undergoes a conformational change due to the fusion peptide attaching to the target cell membrane, exposing the pre-hairpin structure of the HR1 trimer (Figure 3 ). This trimer contains three highly conserved hydrophobic furrows, which then become exposed.
Upon entering the host, the virus binds the transmembrane serine protease 2 (TMPRSS2), leading to the activation of the S protein and formation of the RBD via proteolytic cleavage. The RBD can now interact with the ACE2 receptor, facilitating viral fusion with the membrane and allowing replication within the host.
The hydrophobic furrows formed by the stiff helix and a flexible loop of the HR2 domain facilitate interaction with the HR1 domain. The S protein comprises two structural domains: the core subdomain and the external subdomain. The core is highly conserved (Figure 3 ), containing five β strands arranged in an antiparallel manner, with a disulfide bond linking two of these strands. Meanwhile, the external subdomain is characterized by a loop stabilized through a disulfide bond13, 20.
Since the S1 units are highly variable, the conserved sequence of the S2 subunits offers tremendous therapeutic potential for antiviral drug targets. The HR1 and HR2 heptapeptide repeats interact with peptidomimetics derived from these domains, effectively blocking viral entry by disrupting the six-helix bundle (6HB) structure mainly responsible for membrane fusion. Thus, peptidomimetics may be explored for targeted SARS-CoV-2 therapy18.
Furthermore, in addition to the ACE2 receptor, the virus exploits alternative routes for viral entry into host cells, including L-SIGN, DC-SIGN, TIM1, AXL, C-type lectins, CD147, and glucose-regulated protein (GRP78)19. GRP78 is a chaperone involved in protein homeostasis under normal conditions, but in stressful conditions, such as old age, diabetes, and obesity, it is overexpressed on the cellular membrane and facilitates viral entry, inflammation, apoptosis, and cell signaling7, 9.
Genetic Composition
The genome of SARS-CoV-2 comprises about ten open reading frames (ORFs), including ORF1a and ORF1b. ORF1a and ORF1b encode a total of 16 non-structural proteins (NSPs), namely, nsp1–16, while six accessory proteins are also present. The genome also contains regions that encode for specific viral proteins, including Plpro, 3CLPro, RdRp, Hel, and S, which include the N-terminal domain (NTD), RBD, SD1, SD2, FL, HR1, HR2, and TM. The dotted line in Figure 4 indicates the furin cleavage sites of the S1/S2 and S2' and TMPRSS221, 22, 23, 24.
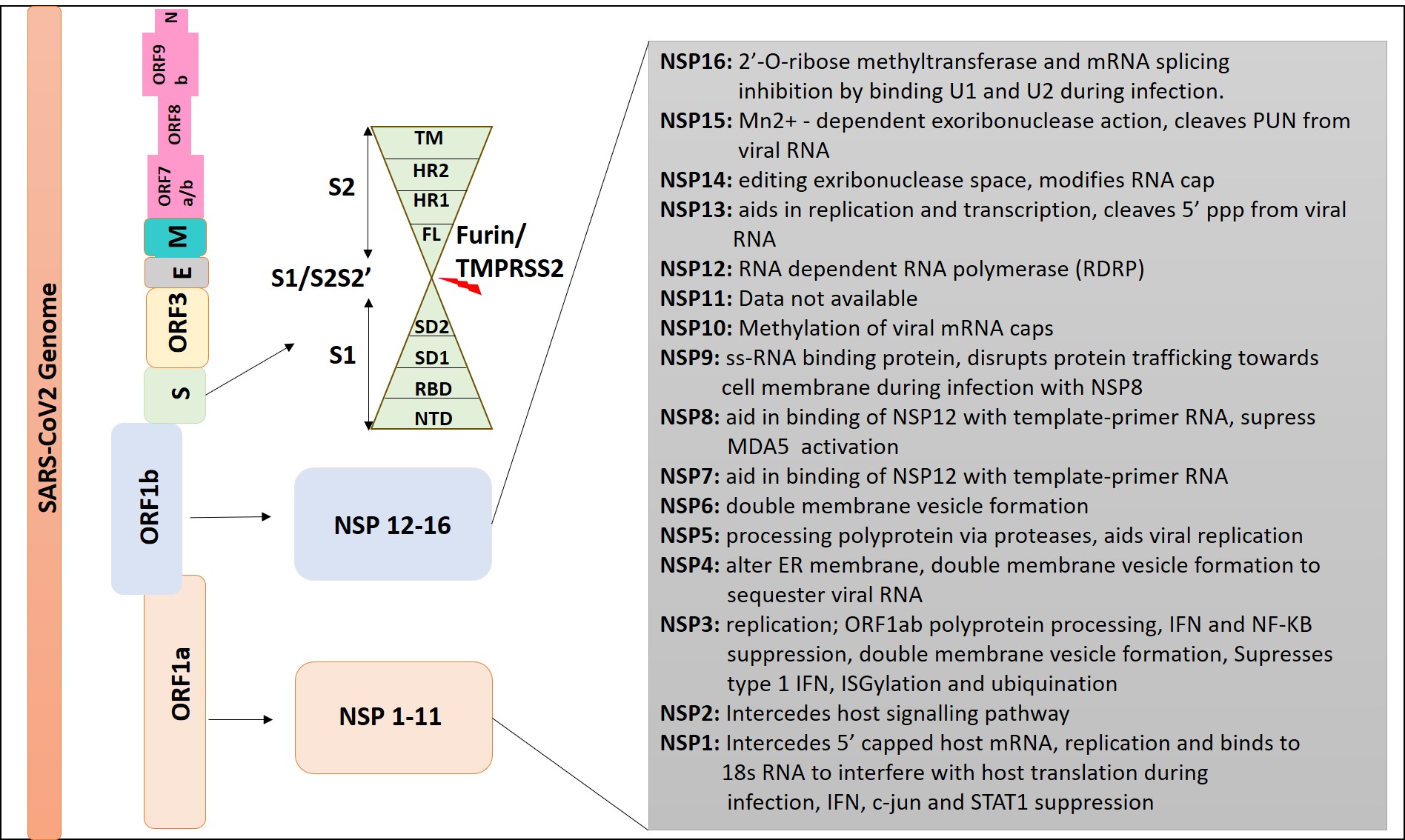
Schematic representation of SARS CoV-2 genome. The viral genome of SARS CoV-2 comprises of ORF1a, ORF1b, S, ORF3, E, M, ORF6, ORF7 (7a and 7b), ORF8, ORF9b, and N sequentially.
Open Reading Frames (ORFs)
The genome of SARS-CoV-2 consists of positive-sense RNA with a 5’ cap and a 3’ poly-A tail, characterized by a low G+C content of approximately 38%. The virus encodes N, E, M, and S structural proteins, which serve as major virulence factors25. SARS-CoV-2 contains the six ORFs that are common to all CoVs. At the 5′ end, two overlapping ORFs, ORF1a and ORF1b, are translated from the genome to generate the continuous polypeptide pp1ab.
About two-thirds of the positive-strand genome is constituted by ORF1a and ORF1b. ORF1a translates into pp1a, which then generates polyproteins pp1a and pp1ab, collectively giving rise to 16 NSPs. NSP3, a papain-like protease (PLpro), facilitates the polypeptide cleavage. NSP5 (Mpro/3CLpro), the primary protease, results in the formation of various non-structural proteins as depicted in Figure 423, 25. Mpro is a significant protease that undertakes the initial and most crucial viral polyprotein cleavages at distinct sites, producing functional proteins. Thus, inhibition of Mpro could be a viable anti-CoV drug target26. The polyprotein pp1a undergoes enzymatic cleavage to yield 11 NSPs, while pp1ab forms 15 NSPs. Further, six auxiliary proteins are encoded by ORFs 3a, 6, 7a, 8, and 10 (Figure 4 )13, 23.
RNA-dependent RNA polymerase (RdRp)
Nsp12 functions as an RdRp, serving as a fundamental functional unit in viral replication and transcription mechanisms. Essential cofactors nsp7 and nsp8 facilitate the binding of RdRp to its template RNA strand and enhance its enzymatic activity. It is essential for replication and transcription machinery of SARS-CoV-2. Structurally, it exhibits a right hand-like shape and possesses domains such as the N-terminal nidovirus RdRp-associated nucleotidyl transferase (NiRAN), the C-terminal region, and an interface domain. This enzyme is a primary focus for many antiviral drugs like remdesivir27, 28.
The 3’ ORF, encompassing the remaining genome (including genes encoding for the remaining structural and auxiliary proteins), is formed via a replication-transcription complex (RTC)22, 23. The viral 3’ untranslated region (UTR) within the genome harbors the initial binding site of the RTC and multiple cis-acting regulatory elements crucial for the viral genome transcription and replication29.
Studies have indicated that beta-coronaviruses adjust the length of the coronaviral poly-A tail during the infection of human rectum tumor 18 (HRT18) cells. Initially ranging from approximately 26–45 nucleotides, the poly-A tail of the viral RNA reaches a peak of about 65 nucleotides between 6–10 hours post-infection. Subsequently, this length gradually decreases to 30–45 nucleotides after 10 hours. Additionally, it has been determined that the effectiveness of virus translation is linked to the length of the coronaviral poly-A tail30. ORF3 hinders host interferon (IFN) signaling and triggers the inflammasome and apoptosis of the host cell. ORF7a also inhibits the host protein translation and promotes pro-inflammatory signaling. ORF N inhibits the RNA sensing mechanisms and STAT1/2, and ORF M suppresses the RIG-1, MDA5, and TRAF-3–TANK–TBK1/IKK epsilon complex31.
Details of the different mutant strains of SARS CoV-2
|
Name of Strain |
Mutation |
Origin of Mutation |
Reference |
|
Gamma |
P.1 |
January 2020 in Japan/Brazil |
|
|
Zeta |
P.2 |
April 2020 in Brazil | |
|
Lambda |
C.37 |
August 2020 in Peru | |
|
Iota |
B.1.526 |
November 2020 in New York | |
|
Alpha |
B.1.1.7 |
December 2020 in USA | |
|
Beta |
B.1.351 |
December 2020 in South Africa | |
|
Eta |
B.1.525 |
December 2020 in UK & Nigeria | |
|
Theta |
P.3 |
February 2021 in Philippines | |
|
Epsilon |
B.1.427 |
March 2021 in USA | |
|
Kapa |
B.1.617.1 |
April 2021 in India | |
|
Delta |
B.1.617.2 |
May 2021in India | |
|
Mu |
B.1.621 |
August 2021 in Columbia | |
|
Omicron |
B.1.1.529 |
November 2021 in South Africa |
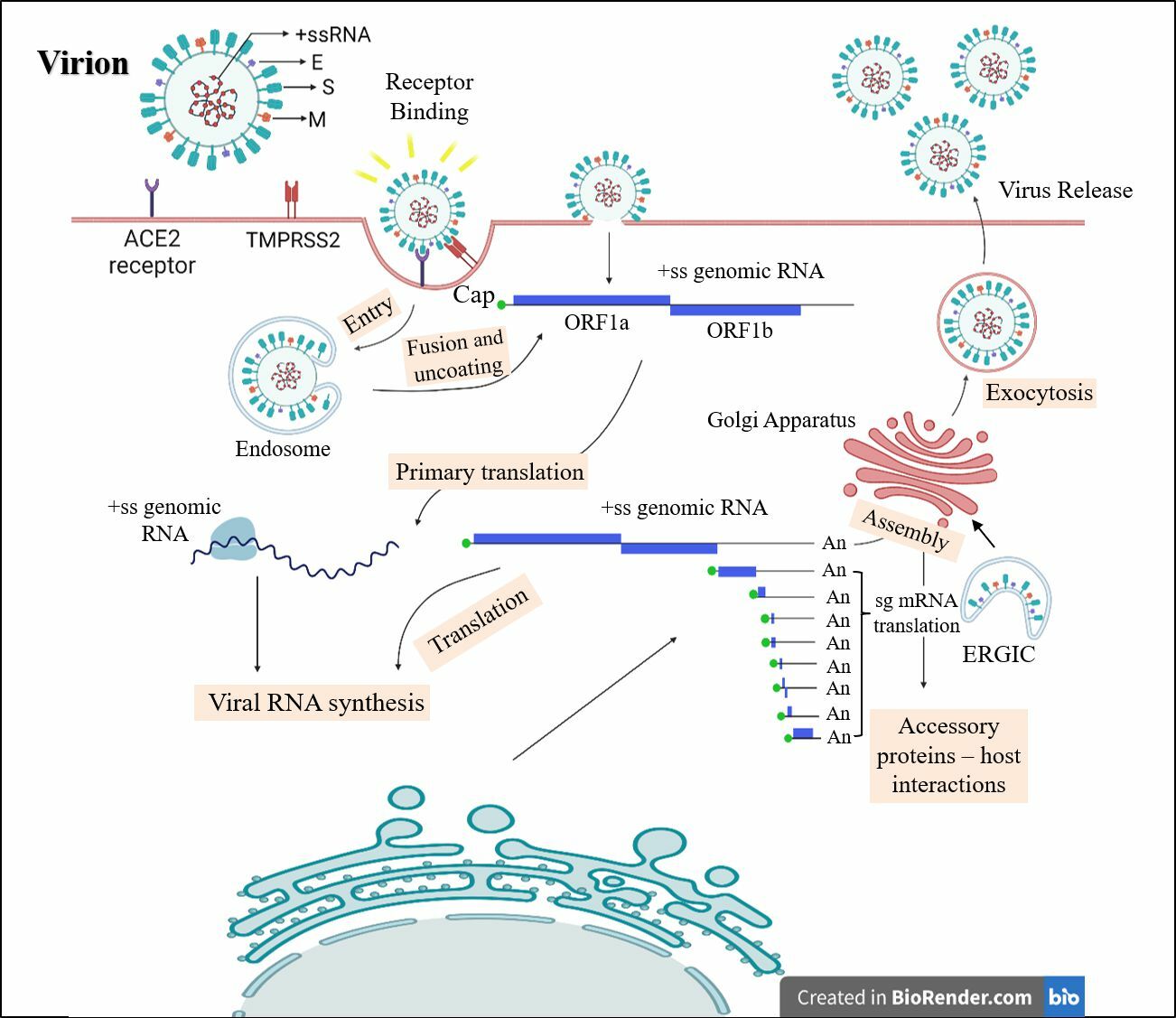
Infection cycle of SARS CoV-2 virus in a human host. SARS CoV-2 characterized by its positive-sense ssRNA (+ ssRNA) genome is encapsidated by nucleocapsid (N) protein and incorporated into the virion by membrane (M) and envelope (E) proteins.
S proteins
The viral S proteins are major virulence factors, and any mutation in the RBD can lead to a new variant with varying virulence levels. Bioinformatics analysis revealed the most probable mutation sites across various SARS-CoV-2 strains (
Consequently, infections caused by BF.7 variant have resulted in significant disease in people with weakened immune systems34. The high transmissibility of the coronavirus and its rapid pandemic spread may be attributed to RNA as a genetic material prone to mutations and the airborne nature of the virus.
The Receptor Binding Domain: The RBD consists of a core made up of beta sheets with antiparallel strands covered with alpha helices and a loop called the receptor binding motif, which wraps around the ACE-2 receptor, thereby initiating direct contact35, 36. The S2 subunit is a transmembrane domain containing fusion peptides involved in the fusion of viral and cellular membranes through conformational modifications32, 37. The RBD also contains three of the missense mutations that are otherwise present in the non-structural and structural proteins, with the exception of the E protein.
High Infectivity of SARS-CoV-2: The virus spike proteins in SARS-CoV-2 show a notably higher infection rate compared to other SARS-CoVs, primarily due to an additional furin cleavage site containing multi-basic amino acids at the junction between the S1 and S2 subunits of the S protein38. The S1 unit is located on the membrane surface and contains the RBD, which specifically binds to the ACE-2 receptor in human hosts, thereby determining the specificity and pathogenicity (37). ACE-2 receptors are widely expressed on epithelial cells of the upper respiratory tract and endothelial cells of arteries, veins, immune cells, and cerebral neurons; this demonstrates the wide range of infectivity of the virus. Thus, in children, the tubular epithelial cells of the kidney, intestinal mucosal cells, and renal epithelial cells become potent targets for viral infection39.
Two prominent strains of the SARS-CoV-2 virus were discovered. According to Tang ., these strains were found by analyzing 103 different virus genome sequences. Less than 30% of these were of type S, while 70% were of type L. The Wuhan, China outbreak began with a high prevalence of the L type; however, this has since declined. Type L is considered to be more aggressive and contagious than type S40.
Upon viral entry in the host cell, it adopts two strategies: first, the E proteins continue the cycle by contributing to viral lysis and ultimately releasing the viral genome into the cell39. The entry of the viral genome marks the beginning of the replication process, which is a complex process involving numerous NSPs23. During this process, multiple copies of the negative sense ss-RNA are created, which act as a template for the synthesis of the positive sense genomic RNA copies. These are further translated to form more NSPs and reverse transcription complexes or are packaged into new virion particles with the help of E proteins37, 41. The N protein aids in its genome protection and packages it into a ribonucleoprotein complex, and further suppresses the immune response of the host cell41. M protein interacts with other structural proteins to induce budding, and plays a significant role in the checkpoint system to ensure proper packaging of the virion particle (Figure 5 )26.
The S protein trimer protrudes from the viral envelope and enables specific binding to cellular receptors, ., ACE2, to promote viral entry and fusion. Upon entry, the viral RNA is translated into two large ORFs. These ORFs are subsequently processed into NSPs that assemble into the viral replication and transcription complex. This complex generates viral replication organelles, including characteristic double-membrane vesicles and convoluted membranes. These structures provide a protected environment for genomic RNA replication and sub genomic mRNA transcription. Newly synthesized structural proteins are translocated into the endoplasmic reticulum membrane, where they interact with the newly produced genomic RNA before budding into secretory vesicles. Ultimately, the mature virions are expelled from the infected cell through exocytosis.
Secondly, transcription in CoVs appears more complicated compared to other positive-sense viruses, leading to premature termination and internal initiation. Apart from replication, in transcription there is a peculiar discontinuous step for sub-genomic messenger RNA (sgmRNA) formation characteristic of Nidovirales, thus creating a nested group of sgmRNA with 5’ and 3’ co-terminal with the genome. The leader sequence confers protection from nsp-1-triggered endonucleolytic degradation of capped messenger RNA, thus leading to efficient accumulation of virus mRNA and its proteins during infection42.
Phases of SARS-CoV-2 Infection
According to several reports, there are three distinct phases of infection for SARSV2:
-
• Stage I is the incubation period, which typically lasts around five days, during which the virus may, in some instances, remain asymptomatic and persist undetected in the host.
-
• Stage II is when the virus becomes detectable and presents mild symptoms such as fever or cough.
-
• Stage III marks the onset of severe complications, including acute respiratory distress, which in critical conditions may lead to death
43 .
The majority of patients experience bilateral ground‑glass opacities on chest CT scans, fever, dry cough, and dyspnea. Pneumonia is among the major complications for COVID‑19 patients, generally characterized by slower recovery and notable spatial heterogeneity. Furthermore, ground‑glass opacities (GGOs), respiratory distress, and cardiac injury are among the serious conditions associated with high mortality43.
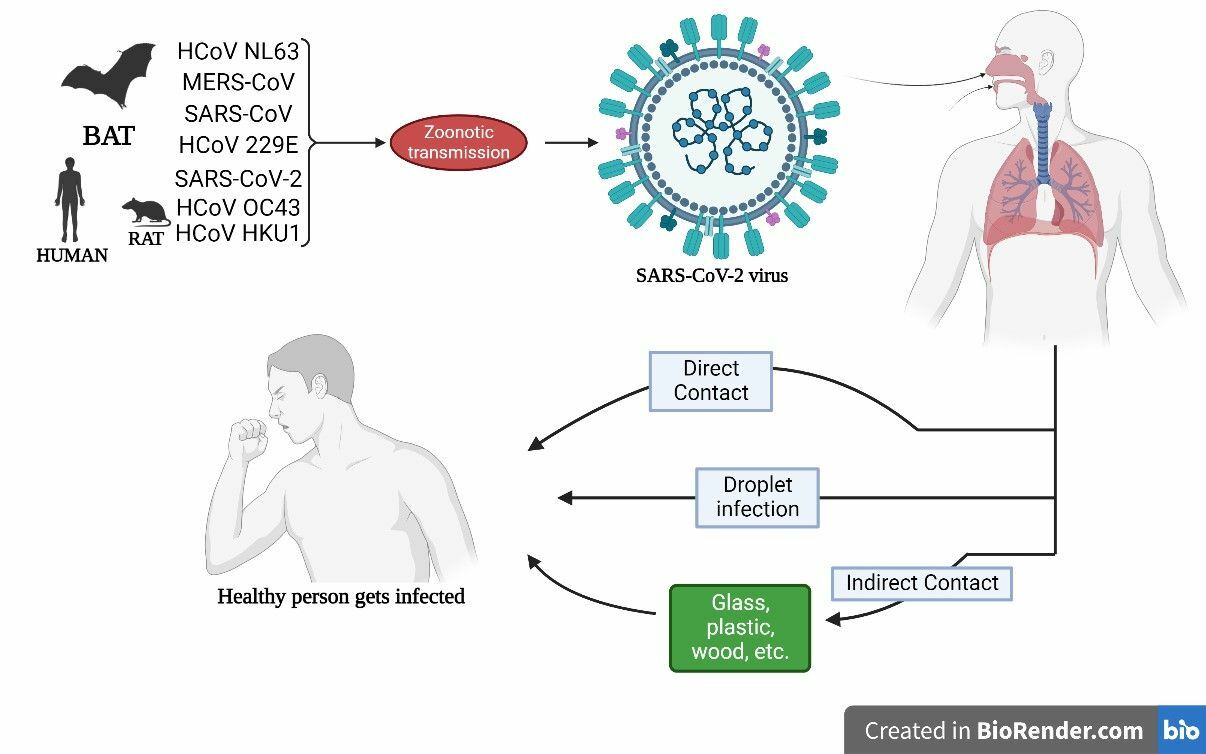
Representation of possible infection cycle and its transmission routes via various modes like direct person-to-person, droplet infection and indirect media-to-person.
Respiratory droplets are the primary airborne mode of transmission; however, the disease can also spread via direct contact with contaminated surfaces and the fecal‑oral route (Figure 6 )44. The reproduction number (R0) and doubling time are estimated to be 2–5 days44. The R0 of the delta variant has significantly increased compared to the parent strain, reaching 5.0845.
The virus eventually decays over time in any environment; however, its half‑life is longest on plastic surfaces, followed by stainless steel46. Chin discovered that SARS‑CoV‑2 is more stable at lower temperatures and becomes dormant after five minutes at 70°C47. Although some studies suggest that SARS-CoV-2 can be transmitted from mother to child via environmental exposure during pregnancy, the possibility of vertical transmission remains controversial, and further research is required in this area48.
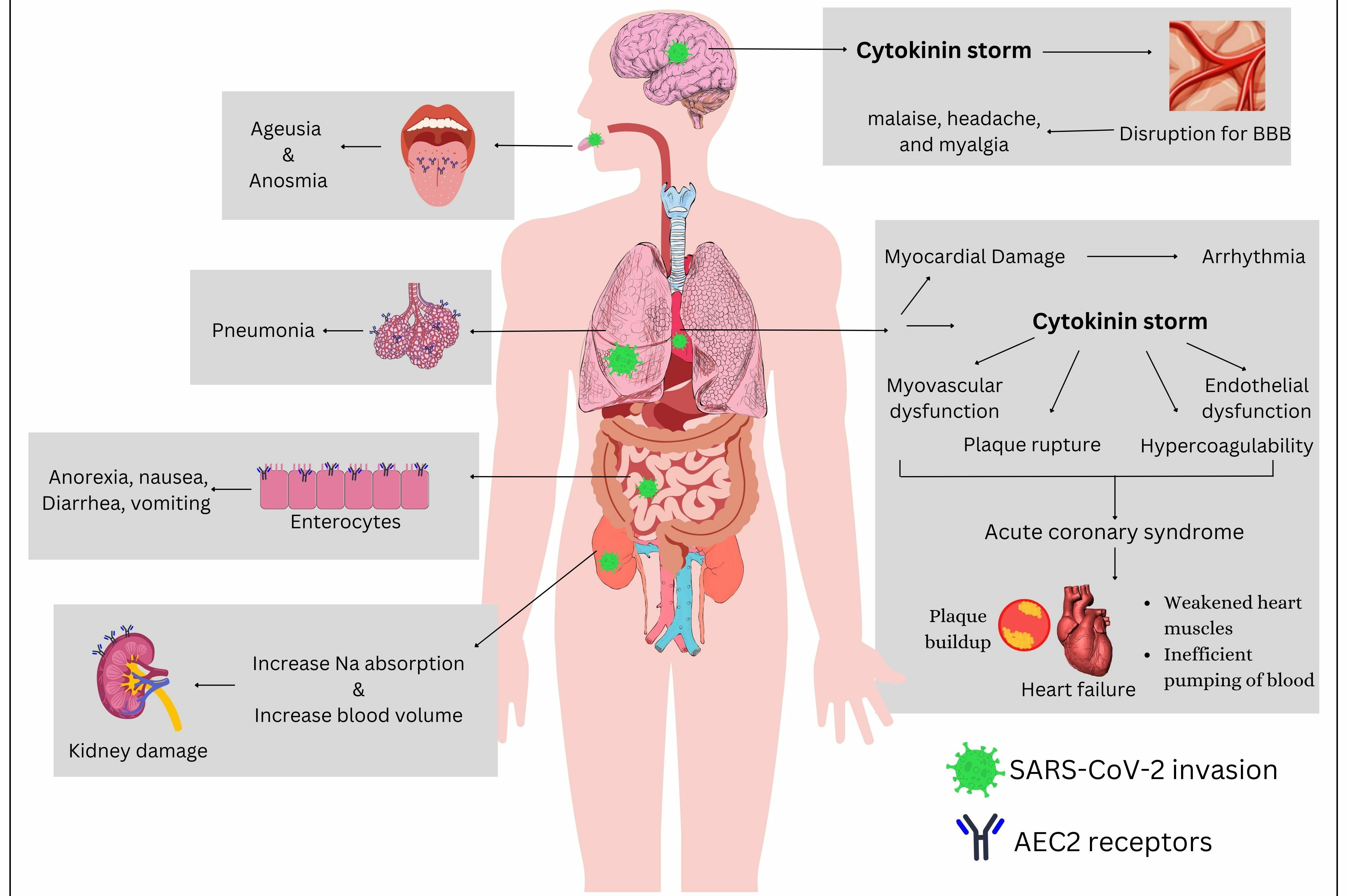
Impact of SARS CoV-2 on various organs and organ systems. SARS CoV-2 can impact multiple organ systems, including the oral cavity, respiratory system, gastrointestinal tract, urinary tract, brain, and cardiovascular system. The virus can accelerate infection and cause organ dysfunction.
Organ Dysfunctions Linked with SARS-CoV-2
The main target of the S protein is the ACE2 receptor, which not only is found in the lungs but is also expressed in various essential organs, including the brain, heart, oral and nasal mucosa, pancreas, gastrointestinal tract, vasculature, and kidneys (Figure 7 ). This broad tissue distribution can lead to multi-organ damage in SARS-CoV-2-infected patients, culminating in multi-organ failure in severe cases35.
Tongue
The most prevalent clinical manifestations of COVID-19 infection are ageusia (loss of taste) and anosmia (loss of smell). Fungiform and surrounding papillae on the tongue contain a significant concentration of ACE2 receptors, which have been implicated in the transient modulation of taste sensitivity through the Renin-Angiotensin System (RAAS) or Renin-Angiotensin System (RAS) enzyme, potentially explaining the loss of taste49. Various sources reveal that SARS-CoV-2 can enter both the peripheral and central nervous systems, leading to detrimental neurological effects50. The virions may infiltrate the olfactory bulb via the cribriform plate of the ethmoid bone from the nose to the brain, causing ageusia and anosmia49. studies in mice have demonstrated the virus’s spread to the brain via the olfactory receptor. Hence, the altered sense of smell can occur due to the virus entering the olfactory bulb, which is responsible for smell perception, through the cribriform plate50, 51, 52.
Nervous System
The neurological symptoms commonly include malaise, headache, and myalgia; in severe cases, ataxia, seizures, altered consciousness, cerebrovascular disease, neuralgia, and low vision may occur35, 53. The detection of SARS-CoV-2 in cerebrospinal fluid (CSF) indicates a potential connection between the virus and the brain. Additionally, the virus is associated with elevated production of cytokines, chemokines, and other immune-modulating molecules, resulting in an intracranial cytokine storm that disrupts the blood-brain barrier49, 50, 54, 55.
Cardiac and Urinary Systems
Patients with cardiac and renal dysfunction are at a higher risk of COVID-associated mortality. Systematic reviews show that common cardiovascular complications in COVID-19 patients include pericarditis, myocarditis, arrhythmias, hypertension, and myocardial infarction56, 57. In cardiac patients, elevated ACE2 receptor levels serve as a protective mechanism against myocardial infarction. However, ACE2 upregulation also increases susceptibility to COVID-19 infection49, 58. Studies indicate that individuals with a history of COVID-19 have a nearly 19–28% higher risk of heart damage, 62% of hospitalized cases experience acute myocardial injuries, and about 33% of these cases result in death56. Emerging evidence suggests that people recovering from COVID-19 face a significantly increased risk of cardiovascular events, including a doubled chance of pulmonary embolism up to 4.5 months post-infection, with heightened risks of myocardial infarction and ischemic stroke persisting for several months afterward59. These heightened risks are associated with prothrombotic responses induced by the virus, viral cytotoxicity, endothelial damage, hyperimmune responses, and vasculitis. Notably, even individuals without prior heart disease have shown subclinical alterations in cardiac function post-COVID-19, suggesting possible long-term myocardial effects56, 57.
Additionally, COVID-19 has been implicated in acute kidney injury, primarily through direct viral invasion of renal cells, thrombic events, and systemic inflammation. Routine renal functions may be compromised by heightened sodium absorption, leading to elevated blood pressure35. Several laboratory results have detected the virus in urine samples. The vasculature regulating nitric oxide levels—crucial for vasodilation—is also under the control of ACE2 receptors. Increased viral load downregulates ACE2 expression on the cell surface, resulting in vascular dysfunction and inflammation. Histopathological analyses often reveal acute tubular necrosis and glomerular injury. In one retrospective cohort study of 372 patients, those with mixed etiologies of acute kidney injury exhibited the highest mortality rates (90.5%) and required dialysis (85.1%). The study emphasizes that the severity and outcome of acute kidney injury in COVID-19 patients (OR= 4.8, CI= 1.7–13.4, p = 0.003) are closely tied to the underlying pathophysiological mechanisms. These findings underscore the critical need for ongoing monitoring of cardiovascular and renal functions in COVID-19 patients, even months after the acute infection phase60, 61.
Recently, additional symptoms have been documented in some patients, primarily affecting the fingers and toes, referred to as ‘COVID toes.’ These red and purple painful lesions tend to appear in later stages and could be linked to vascular complications62, 63.
Comorbidities associated with SARS CoV-2
Diabetes
Globally, diabetic patients are more susceptible to SARS and MERS infection, potentially worsening the condition. Factors contributing to this include compromised immunity due to reduced phagocytic activity, lower neutrophil function, and decreased T-cell performance. Diabetes results in elevated ACE2 receptor expression, leading to enhanced viral affinity and increased viral load64 Wijnant reported higher bronchial and alveolar ACE2 expression in patients with diabetes, independent of other factors such as smoking, body mass index, comorbidities, or the usage of RAAS inhibitors. Furthermore, a direct association has been observed between blood glucose levels and alveolar ACE2 expression, which supports viral replication and may lead to potentially fatal complications due to immune system dysregulation and an exacerbated inflammatory response64, 65.
Cancer
Cancer patients, being immunocompromised, are at a heightened risk for SARS-CoV-2 infection, which can increase disease severity and mortality41, 66, 67. COVID-19-positive lung cancer patients demonstrated higher inpatient mortality. Prolonged prothrombin expression and increased Troponin I sensitivity are distinct prognostic factors, helping physicians determine outcomes earlier. These patients often present atypical early symptoms and distinct imaging findings68.
Those with hematologic cancer, lung cancer, and other malignancies in advanced stages have shown a greater incidence of serious events related to SARS-CoV-2, including elevated mortality and severe symptoms69. Breast and gynecologic malignancies were found in around 50% of cancer patients examined for SARS-CoV-2 seroprevalence, suggesting an increased infection rate among females.
In a study by Cury ., COVID-19 interactions with lung cancer were examined in A549 and Calu-3 cell lines, revealing overexpression of the cancer-linked genes BRCA1 and CENPF in small and non-small cell lung cancer, respectively, which may interact with the viral S protein and helicase (NSP13). No changes in ACE2 were observed in either cell line, but type I interferons induced ACE2 overexpression during host immune responses. The expression of ACE2 and TMPRSS2 in patients with lung adenocarcinoma and squamous cell carcinoma could suggest an increased susceptibility to infection70.
Asthma
Asthma is a complex respiratory illness characterized by various inflammatory processes. Notably, standard asthma treatment involves corticosteroids and anti-asthmatic medications, which may act as potential modulators of SARS-CoV-2 infection, potentially lowering its prevalence in individuals with asthma71.
Human Immunodeficiency Virus (HIV)
Ssentongo . reported an increased risk of infection and higher mortality in individuals with both COVID-19 and HIV. HIV-induced immunosuppression elevates the risk of additional comorbidities such as anemia, neutropenia, and thrombocytopenia, which can hinder COVID-19 recovery. Therefore, coinfection with COVID-19 and HIV can lead to serious outcomes if not managed effectively72.
A recent comparative study demonstrated that HIV patients with COVID-19 experience delayed recovery and lymphocytopenia, primarily attributed to reduced SARS CoV-2-specific antibody responses41.
Clinical evidence suggests that HIV-positive individuals on antiretroviral therapy exhibit reduced vulnerability to infection. However, HIV-related chronic conditions, a low CD4 (<200 cells) cell count, and uncontrolled HIV viremia have been associated with worsened COVID-19 severity and increased mortality73.
Pregnancy
Viral infections can alter the mother’s innate immune response during pregnancy74. In the first trimester, increased ACE2 receptor expression is observed on villous cytotrophoblasts, syncytiotrophoblasts, decidual cells, and the placenta, making pregnant women more susceptible and raising the possibility of vertical transmission. The blastocyst and embryos (of good quality and euploid) were also affected75.
The effect of SARS-CoV-2 on female fertility has been evaluated, showing no significant impact on ovarian reserves. Regarding the menstrual cycle, enhanced variability was noted in mild to severe infections, but these changes were reversible. Conversely, the virus may significantly affect the sexual and pubertal development, as well as fertility in young male children, by disrupting the neuroendocrine axis. In adult males, SARS-CoV-2 infection could impede spermatogenesis via considerable damage to testicular tissue, particularly impacting Sertoli cells76.
Long COVID-19: Post-Acute Effects
In accordance with the WHO, the persistence of COVID-19 symptoms such as muscle and joint pain, fatigue, diarrhea, abdominal pain, and cough for at least three months following recovery, and continuing for at least two months without an alternative diagnosis, is referred to as Long COVID-1977. It affects multiple organ systems, leading to chronic fatigue, respiratory issues, neurological complications, cardiovascular problems, and mental health disorders. Long COVID may arise from persistent viral reservoirs in certain organs after the primary infection, which repeatedly trigger the immune system and lead to chronic inflammation. In some patients, autoantibodies are produced, attacking their own tissues. Furthermore, microclots, impaired circulation, and vascular damage during the initial infection can reduce oxygen delivery, contributing to fatigue, organ damage, and cognitive dysfunction in Long COVID patients78.
According to a meta-analysis by Notarte (2022), older age (OR 0.86, p = 0.17) may not be associated with the sequelae of Long COVID-19. However, being female (OR = 1.48, p = 0.01), having diabetes, pulmonary disease, a transplant history, and obesity may serve as risk factors79. Furthermore, a comparative meta-analysis examined post-COVID-19 cases to compare the prevalence of symptoms in both non-hospitalized and hospitalized patients. Dyspnea and fatigue (35–60%), cough (20–25%), ageusia (15–20%), joint pain (15–20%), and anosmia (10–20%) were documented, appearing in ≥60% of the cases80.
In 2022, César Fernández-de-las-Peñas . evaluated the prevalence of Long COVID-19 in individuals with the original viral strain and compared it to those infected by the Alpha, Omicron, and Delta variants. The original strain group experienced 50% more episodes of Long COVID-19, with fatigue being the most frequently reported symptom80.
Although COVID-19 vaccination lowers the risk of severe disease, its effect on Long COVID-19 outcomes was initially unclear. A systematic review by Notarte . (2022) indicated that receiving two vaccine doses correlated with reduced Long COVID-19 cases. Individuals who had at least one month between vaccination and COVID-19 infection were less likely to experience Long COVID-19 sequelae81, 82. Additional studies suggest that vaccinated individuals have a lower risk of developing Long COVID, likely due to reduced viral replication, systemic inflammation, and persistent symptoms83. Moreover, those vaccinated after infection often recover more quickly from Long COVID, as vaccination modulates the immune response and prevents the inflammation associated with prolonged symptoms. Long COVID frequently causes fatigue and neurological complications; by limiting viral spread to the nervous system, vaccination reduces these risks. Therefore, vaccines play a crucial role in decreasing disease severity and the risk of Long COVID84.
Age
Age is among the most significant predictors of COVID-19 severity and mortality. Individuals over 60, particularly those over 70, have markedly higher hospitalization and mortality rates. Furthermore, the immune system weakens with age—a process known as immunosenescence—reducing the body’s ability to combat infections. Older adults are also more likely to have comorbidities such as heart disease, diabetes, and hypertension, which can worsen COVID-19 outcomes85.
By contrast, children and young adults typically experience milder symptoms or remain asymptomatic, though they can serve as asymptomatic carriers, transmitting the virus to more susceptible groups. Additionally, a small proportion of children develop multisystem inflammatory syndrome (MIS-C), a rare but serious COVID-19 complication86.
Gender
COVID-19 outcomes vary between males and females because of both biological and behavioral factors. Men often experience more severe disease and higher mortality rates, partly due to weaker innate and adaptive immune responses and lifestyle factors such as higher rates of smoking and alcohol consumption, which contribute to immune suppression and lung damage. Women, on the other hand, benefit from stronger immune mechanisms influenced by genetic factors and estrogen. However, pregnant individuals face heightened risks and complications from COVID-19, as previously discussed87.
Socioeconomic Status
Socioeconomic inequalities profoundly affect COVID-19 outcomes by influencing infection rates, healthcare access, and mortality. Individuals in low-income communities often face limited healthcare accessibility, resulting in delayed diagnoses and treatments. Lower vaccination rates also increase vulnerability to severe illness. Additionally, many frontline workers (., healthcare and transportation staff) have higher exposure risks. Overcrowded living conditions and shared facilities in lower-income settings make social distancing difficult, further raising transmission risks. Poor nutrition and inadequate healthcare compound these problems88, 89.
A systematic review and meta-analysis of 72 studies—mostly from the United States—found that Black individuals experienced significantly higher COVID-19 infection (156%), hospitalization (153%), and mortality (105%) rates than white individuals90. In England, a study using a Bayesian hierarchical spatio-temporal model showed that areas with lower household incomes and higher unemployment rates had elevated COVID-19 infection rates. Moreover, a UK report found that residents of the most deprived areas were nearly twice as likely to be hospitalized for infectious diseases, including COVID-19, compared to those in wealthier regions. These findings shed light on the critical impact of socioeconomic factors on COVID-19 outcomes91.
Race and Ethnicity
In the United States, African American, non-Hispanic, and Indigenous populations show higher COVID-19 hospitalization and mortality rates. These disparities often result from a combination of genetic, socioeconomic, and healthcare access factors92, 93. A broad analysis of national data revealed that marginalized communities, including Black, Hispanic, and Native American individuals, faced more than double the hospitalization rate compared to White patients; age-adjusted mortality rates were also highest among these groups. Additionally, a multi-hospital study of critically ill COVID-19 patients found an increased in-hospital death risk for Hispanic patients relative to non-Hispanic patients. Such differences highlight the interplay of socioeconomic status, healthcare accessibility, and potential genetic predispositions94, 95.
Research from India and Bangladesh indicates that people with preexisting conditions such as hypertension, diabetes, and asthma are at higher risk for extended post-COVID-19 symptoms. A South Indian study reported that 39.6% of patients continued experiencing Long COVID symptoms past 16 weeks post-infection, with older age, hypertension, and asthma identified as significant predictors. In Bangladesh, more than 62% of hospitalized COVID-19 patients had comorbidities, which prolonged hospital stays and increased post-discharge complications96, 97. These findings emphasize the need for tailored healthcare strategies to address the requirements of patients with multiple health conditions.
Diagnostic Techniques
Diagnostics are absolutely critical and irreplaceable for combating any highly contagious illness. Because of the SARS CoV-2 emergency, the world experienced a significant shift and advancement in diagnostic practices98. However, the urgent need to expand diagnostic capacity to meet growing demands led to the recruitment of staff lacking essential technical expertise, notably in specimen processing, troubleshooting, and result/error analysis. Additionally, issues such as low test sensitivity, timing challenges, low viral loads, and unvalidated kits exacerbated these problems99. Such diagnostic errors can elevate the risk of cross-contamination, raise the incidence of false-positive or false-negative results, and undermine the effectiveness of health policies, surveillance, and containment strategies100. Therefore, early identification of SARS CoV-2 is vital for halting its spread, especially given its asymptomatic phase. Diagnosis can be achieved through immunological methods (ELISA, Rapid Antigen Test, Liver Function Test) or molecular techniques (RT-PCR, LAMP).
Immunological tests detect antigens (Ag) or antibodies (Ab) in the lungs and blood using serological assays, helping us understand disease transmission and its underlying mechanisms. Molecular diagnostics, by contrast, target nucleic acid traces, enabling earlier pathogen detection, which is crucial for virus containment. Moreover, non-invasive approaches like CT scans and X-rays contribute to early disease detection101. While several methodologies are well-established, numerous innovative diagnostic tools have been developed and refined to identify the infectious agent, and many remain under investigation98 (Figure 8 ).
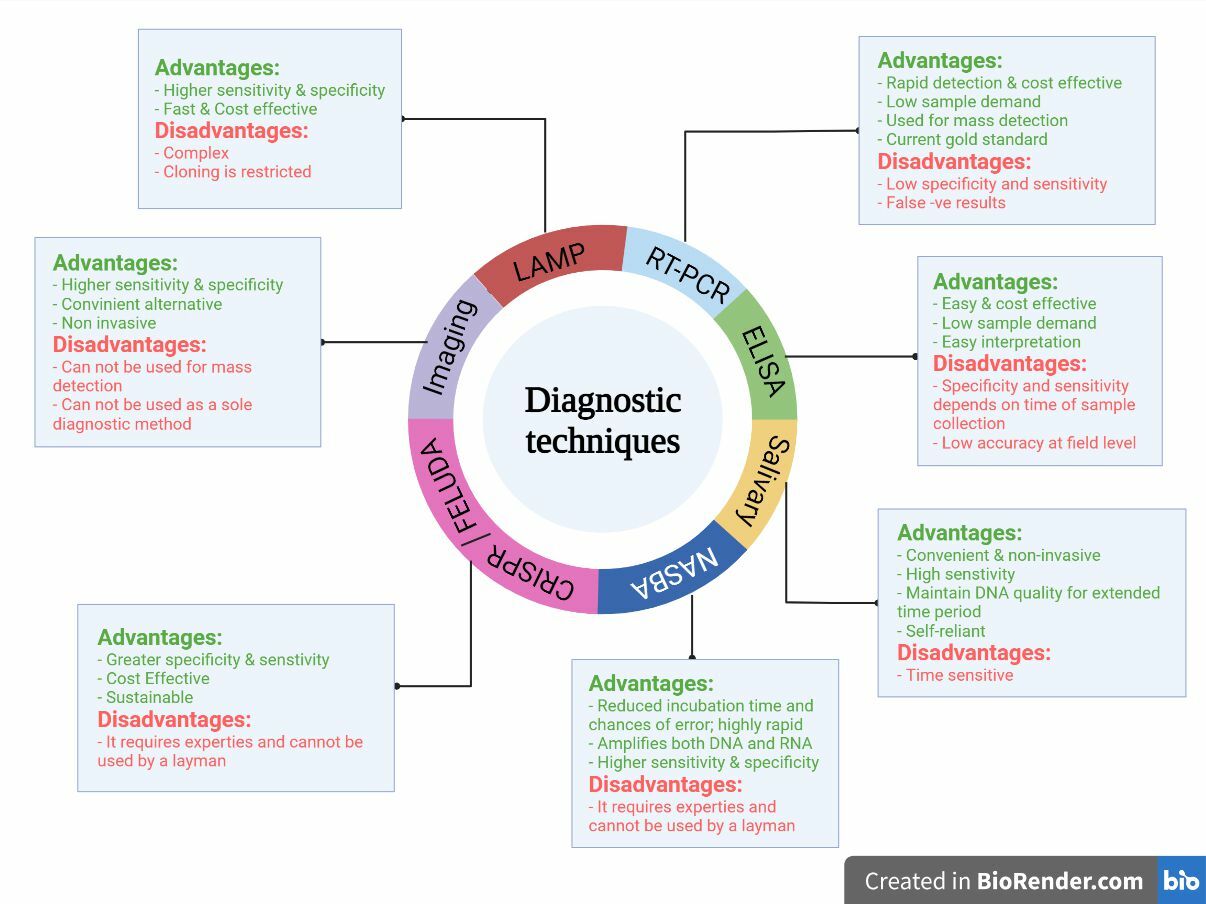
Advantages and disadvantages of various diagnostic tools and techniques for detection of COVID-19.
Polymerase Chain Reaction (PCR)
Real-time polymerase chain reaction (RT-PCR) has been widely recognized as the gold standard for diagnosing COVID-19 due to its high specificity and reliability. This rapid detection method can detect and amplify even a few copies of the viral genome, making it an ideal diagnostic approach.
However, RT-PCR has several limitations, including false-negative results stemming from improper sample collection. Transport and storage conditions are critical for ensuring accurate results, as any deviation may compromise test precision. Moreover, the timing of testing is crucial because viral titer varies during infection; testing too early or late may result in undetectable virus. Mutations in the viral RNA can affect primer binding and reduce test sensitivity. Additionally, RT-PCR can detect viral RNA even after patient recovery, leading to unnecessary extended isolation. The method also requires expensive equipment, skilled personnel, and specialized settings, restricting its use in low-resource laboratories102.
Despite these limitations, RT-PCR remains an effective method to confirm infection when common symptoms such as cold, cough, fever, joint pains, or loss of taste and smell are present. Because gene-specific primers target the E or N proteins of the virus, this technique delivers accurate positive results and minimizes the risk of detecting other strains103. Many studies worldwide aim to develop novel assays that target various viral markers, including the S protein, N protein, and RdRp/helicase genes, in order to enhance the efficiency of RT-PCR compared to conventional assays, which primarily probe the N protein and ORF1b region104.
Nevertheless, some samples that tested negative in early RT-PCR were later confirmed positive, indicating that a negative result cannot guarantee the absence of SARS-CoV-2103, 105. Genetic mutations at primer or probe binding sites may contribute to false results. Although RT-PCR offers a relatively cost- and time-effective solution, it still does not achieve 100% specificity or sensitivity103.
Fang . (2020) investigated a small cohort (n=51) to compare lung CT and RT-PCR for diagnosing COVID-19 (106). Following an average of ±3 days from disease onset, the first RT-PCR round identified 36/51 positive cases, with 12, 7, and 1 confirmed by 2, 3, and 4 subsequent assays, respectively. In contrast, lung CT scans detected 50/51 positives, demonstrating a 98% specificity compared to 71% for RT-PCR. Hence, negative RT-PCR results may require additional confirmation106.
Digital PCR (dPCR), particularly droplet digital PCR, has emerged as a robust tool for highly precise quantification of viral RNA. Its exceptional sensitivity supports the detection of low viral loads, making it invaluable for identifying SARS-CoV-2 variants, including Spike protein mutants (R346T, N460K, K444T, F486S, F486V) and Omicron subvariants, in clinical samples. These assays can detect up to four single nucleotide polymorphisms (SNPs) simultaneously, enabling accurate discrimination among subvariants (BA.2.75.2, BQ.1, XXB)107. Furthermore, Sciensano has developed multiplex droplet digital PCR for SARS-CoV-2 and variant detection in both human and environmental samples108. Overall, the integration of dPCR technologies and multiplex assays has significantly enhanced the capacity for precise, efficient SARS-CoV-2 variant detection, strengthening public health measures and informing targeted interventions.
Enzyme-Linked Immunosorbent Assay (ELISA)
ELISA is an antibody-based detection technique used to detect SARS-CoV-2 in infected patients. The specificity and sensitivity of the technique are highly dependent on the time of testing, as the seroconversion of antibodies primarily depends on the onset of symptoms109.
It is a plate-based assay in which the microtiter wells are coated with viral antigens. After the sample is introduced, the antigen-specific antibodies bind to these antigens. Following a wash step, a conjugate and substrate are added sequentially. The conjugate first binds to the antigen–antibody complex, while the substrate causes a color change upon reacting with the conjugate. The intensity of this color change is directly proportional to the concentration of antibodies in the sample110.
To simplify the process, diagnostic companies usually provide pre-coated ELISA kits labeled with either Ag or Ab for convenient detection of antibodies against the viral genome 109. Antigen tests are widely used for rapid detection but face critical shortcomings, including lower sensitivity than RT-PCR, particularly in asymptomatic or early infection cases. They also yield higher false negative rates, leading to undetected infections in infected individuals. Furthermore, accuracy can vary among different brands, and sensitivity is directly associated with viral titer in the sample, which may fluctuate over time. Cross-reactivity with other coronaviruses can again produce false positives, and in resource-limited settings a positive result often requires confirmatory RT-PCR testing109.
Salivary Diagnosis
Saliva is a convenient and non-invasive option for diagnosis. The virus is present in the saliva of COVID-19-positive patients for up to several days of hospitalization and infection. Studies indicate the presence of active viral particles in the salivary sample for an average of 18–20 days in both mild and severe cases111, 112. Hence, it is a simple approach that should be further validated as a safe option where patients with low sputum levels can also collect their own samples, thus reducing risks to healthcare workers. The field of salivaomics can be further explored and can play a pivotal role in COVID-19 diagnosis through mass-scale screening112, 113.
Nucleic Acid Sequencing Based Amplification (NASBA)
NASBA is a two-step amplification process wherein the genetic material is first denatured and then undergoes a polymerase-dependent amplification cycle. The process is carried out under isothermal conditions with the addition of fluorochromes to make real-time observations98. This technique employs reverse transcription and T7 RNA polymerase, which rapidly amplifies the RNA of interest. The technique offers various advantages over gold standards like RT-PCR. It is a rapid detection method that does not require additional equipment like thermocyclers and can amplify both DNA and RNA effectively with the help of two primers, wherein the amplicons are ssRNA114, 115. The assay is capable of producing billions of genomic copies within 1.5–2 hours, significantly reducing incubation time and minimizing errors. This was used in earlier epidemics to detect SARS-CoV; it is a robust method that is highly sensitive and specific for ssRNA genome diagnosis and has significant potential to be used at the point-of-care in areas with limited resources115. The process has been further modified into RT-NASBA, which provides simultaneous diagnosis of various viral strains. RT-NASBA is faster than RT-PCR and offers 10–100 times higher sensitivity because of the isothermal conditions, where the sample does not require repeated heating or cooling. Thus, this technique holds immense potential and can be employed in the successful diagnosis of SARS-CoV-298.
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and FELUDA for SARS-CoV-2 diagnosis
The CRISPR/Cas system confers acquired immunity in archaeal and bacterial cells by cleaving the DNA or RNA of viral particles, thus rendering them inactive. Because it can cleave both ds and ss RNA and DNA molecules101, 116, this technology can be used for both therapy and diagnosis of COVID-19. It works by degrading the viral ssRNA genome through crRNAs targeting enzymes such as replicase–transcriptase and the S glycoprotein116. This removal of the viral gene prevents the virus from replicating its genome117. Notably, the cleavage activity of Cas13d is independent of PAM-like sequences, enhancing its targeting potential against rapidly mutating viruses116118. Among the Cas proteins, Cas12a and Cas13a appear to be the most effective for diagnostic purposes: Cas12a cleaves DNA, while Cas13a primarily cleaves RNA, making it particularly suitable for virus detection118. Cas12a or Cas13-based diagnostic tools operate under the "collateral cleavage activity" theory (Figure 9). In collateral (or trans) cleavage, Cas12a/Cas13 nucleases, once activated by CRISPR-RNA cleavage (crRNA), non-specifically digest neighboring ssDNA/RNA molecules. This can be observed by performing a lateral flow test on a paper strip with fluorescently labeled ssDNA/RNA reporter probes116.
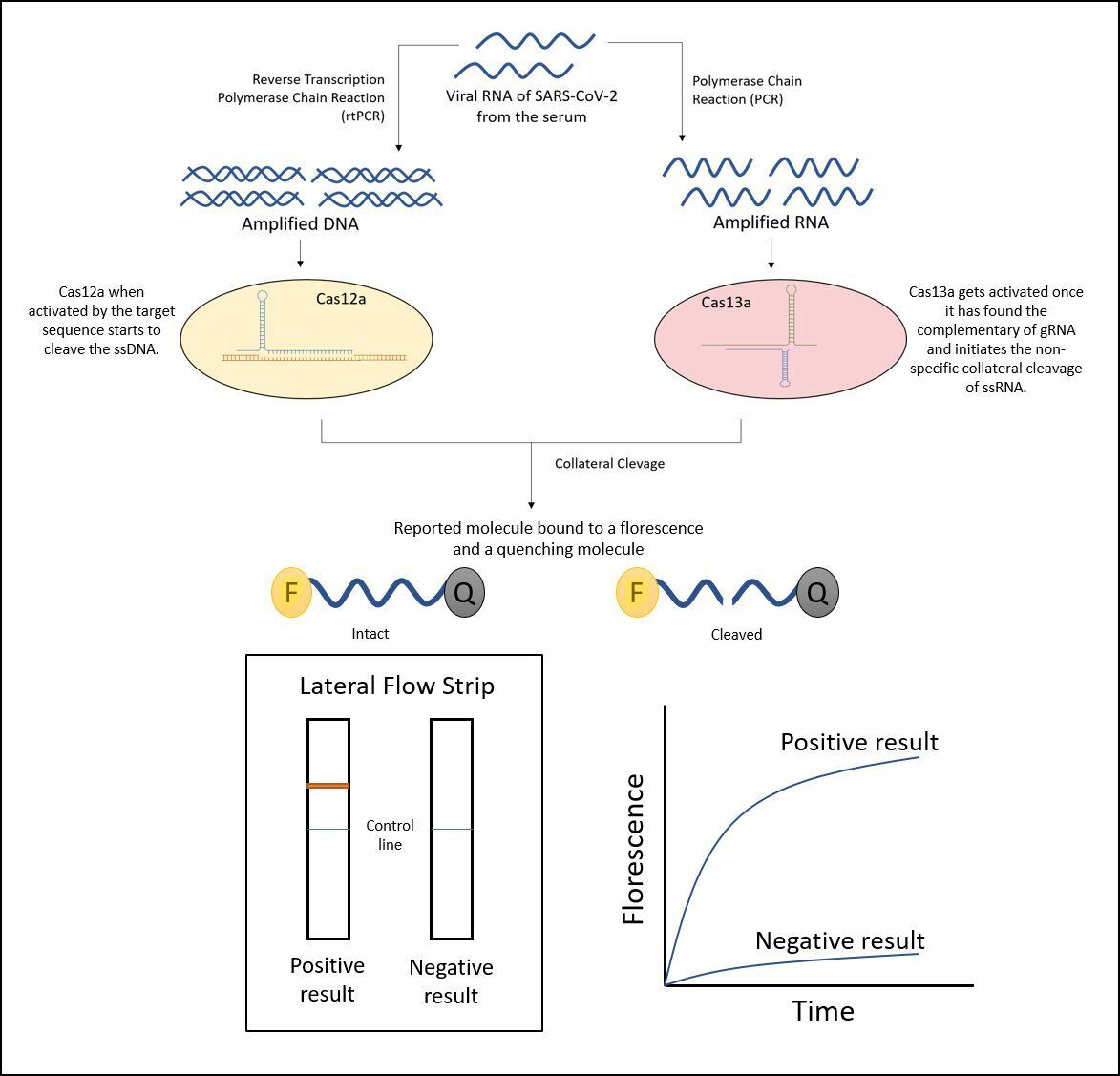
Nucleic Acid Detection of SARS CoV-2 Using CRISPR/Cas Assays.
As a result, Zhang , 2022 introduced a novel technology called SHERLOCK. The test, similarly to qRT-PCR assays, uses read-out RNA extracted from patient samples in under an hour via a dipstick method, eliminating the need for costly equipment17, 118. FELUDA is another promising diagnostic method for COVID-19 detection that is based on CRISPR. This technique is suitable for large-scale diagnostics and can accurately genotype single nucleotide variants, offering other uses in disease diagnostics. When FELUDA is coupled with microfluidics-based fragment analyzers and portable hand-held PCR machines, it enables true point-of-care operations (Figure 9 )119.
The patient’s sample is used to isolate RNA. Both DNA and RNA must then be amplified from the nucleic acid extraction. Cas12a and Cas13 proteins bind to the targeted sequences, initiating a trans cleavage reaction that cuts a fluorophore-labeled ssDNA or RNA sequence, respectively. The cleavage of the fluorophore results in fluorescence, which can be detected using a lateral-flow readout strip, allowing for rapid detection of the virus.
Thus, CRISPR-Cas-based detection methods may supersede the current gold standard (PCR) due to their sensitivity and specificity matching PCR, while delivering highly accurate results. In addition, anticipated costs are low because no specialized or expensive equipment is necessary116. However, CRISPR-based diagnostics often require nucleic acid extraction and preamplifications before detection; therefore, for early stages of infection, CRISPR needs higher concentrations of RNA, making it less sensitive. The need for pre-amplification, RNA extraction, and preprocessing also adds to the cost and turnaround time. Moreover, integration into point-of-care settings is challenging due to multiple reaction steps. Widespread deployment requires specialized reagents and trained personnel, restricting its use in low-resource settings. With the emergence of variants, frequent redesign of guide RNAs is mandatory120. Although anticipated as low-cost and suitable for low-resource environments, widespread adoption is currently limited by the availability and expense of proprietary fluorophore-labeled reporter molecules. Maintaining a reliable cold chain and scaling reagent production for decentralized locations also presents logistical barriers. As CRISPR-based diagnostics are relatively new, their regulatory validation and quality control infrastructure remain underdeveloped in many countries, delaying approvals and limiting field implementations121, 122.
Imaging Techniques: Computed Tomography (CT) and Chest X-Ray (CXR)
One of the earliest and most common imaging tools for detecting pneumonia-related illnesses is chest computed tomography (CT). Ground-glass opacities (GGOs) and consolidation are common CT findings, often near the lung periphery, although a small percentage of individuals exhibit diffuse patterns. Lomoro recently evaluated the CT findings from numerous publications, noting that abnormalities were often bilateral, peripheral, subpleural, and involved the lower lobes123. Previously, it was routinely utilized to detect lung abnormalities in SARS and MERS, and although it may not always detect changes at the onset of COVID-19, it has been found to be more sensitive than X-rays124, 125. Recently, this approach has also been used in hospitals to diagnose COVID-19; however, it has several drawbacks. COVID-19 lung anomalies can mimic those of other bacterial or viral infections, resulting in ineffective detection of the virus alone, but it may prove useful when combined with other techniques such as RT-PCR and may aid in monitoring the progression of COVID-19 infection in the lungs125. Imaging alone cannot differentiate between COVID-19 and other respiratory illnesses.
CXR, similar to CT, also commonly presents increased prevalence in the lower lobes and peripheral regions for most patients. Most individuals experience bilateral involvement. Although these findings are common in infected individuals, they are not specific and can also be observed in other illnesses, including viral and bacterial pneumonia123. However, current imaging techniques often lack specificity; therefore, efforts are being made to enhance the technology in order to provide more accurate diagnoses. Additionally, imaging is costly and requires specialized facilities, limiting its accessibility in resource-limited settings.
Artificial intelligence (AI) can augment traditional radiograph interpretation by employing deep learning techniques to enable screening, triaging, and faster diagnoses. AI might also help track disease progression and identify high-risk patients. In differentiating the CTs of COVID-19 patients from those of other types of pneumonia, Bai and colleagues reported improved accuracy, sensitivity, and specificity. AI can assist radiologists in making the final assessment. In locations where radiologists are limited, AI might be employed to reach the final diagnosis123.
Loop-Mediated Isothermal Amplification (LAMP)
LAMP is based on isothermal nucleic acid amplification for the diagnosis of SARS-CoV-2. It employs six distinct target sequences detected by unique primers in the same reaction, demonstrating high specificity and sensitivity. This faster assay eliminates the need for costly chemicals or equipment, making coronavirus detection more cost-effective126(
Primer sequences used in currently enhanced LAMP assays designed by Lamb
|
Primers |
|
Sequence (5’-3’) |
|
Outer Forward Primer |
F3 |
TCCAGATGAGGATGAAGAAGA |
|
Outer Backward Primer |
B3 |
AGTCTGAACAACTGGTGTAAG |
|
Forward Inner Primer |
FIP(F1c+F2) |
AGAGCAGCAGAAGTGGCACAGGTGATTGTGAAGAAGAAGAG |
|
Backward Inner Primer |
BIP(B1c+B2) |
TCAACCTGAAGAAGAGCAAGAACTGATTGTCCTCACTGCC |
|
Loop Forward Primer |
LoopF |
CTCATATTGAGTTGATGGCTCA |
|
Loop Backwar d Primer |
LoopB |
ACAAACTGTTGGTCAACAAGAC |
The RT-LAMP primers demonstrated a 0% mismatch with the hCoV strains, indicating that these primers would detect all 27 COVID-19 strains examined by Lamp. Furthermore, other CoVs exhibited a mismatch of 27–54% nucleotides with RT-LAMP primers, making it highly unlikely that they would produce a positive RT-LAMP result, thereby confirming the assay's specificity for COVID-19127. However, LAMP is prone to non-specific amplification due to its highly sensitive enzymatic reaction, increasing the likelihood of false positives. Carryover contamination can also lead to erroneous results. Furthermore, at low viral loads (., in early stages of infection or in asymptomatic individuals), LAMP may produce false negatives. With LAMP, 1–2 genes can be targeted; however, multiplexing and variant detection remain challenging128, 129.
Diagnostic Challenges in Long COVID
Long COVID lacks any standardized diagnostic criteria and is primarily a clinical diagnosis, as no single test can confirm it. The variability in its symptoms makes diagnosis challenging. Symptoms such as fatigue and cognitive impairments make it difficult to differentiate long COVID from pre-existing conditions such as depression or anxiety. Laboratory tests and imaging may not always reveal abnormalities despite persistent symptoms. Furthermore, variability in disease presentation further complicates diagnosis. Some patients recover within weeks, while others have relapsing-remitting symptoms for months130.
Vaccination against COVID-19
Vaccination against COVID-19 has played a crucial role in reducing severe disease, hospitalization, and mortality. Studies have shown that vaccinated individuals with comorbidities have a lower risk of severe complications than unvaccinated individuals with similar health conditions. Vaccination also reduces the risk of COVID-19-related cardiac events and strokes by minimizing systemic inflammation131. However, emerging SARS-CoV-2 variants, notably BA.2.86 and its descendant JN.1 (L455S, F456S in the RBD region), have exhibited significant immune evasion capabilities, challenging the efficacy of existing vaccines and therapeutic antibodies. BA.2.86, characterized by over 30 spike protein mutations compared to earlier variants, demonstrates substantial resistance to neutralization. Studies indicate a 57-fold reduction in neutralization efficiency against BA.2.86 compared to the original Wuhan strain in plasma from individuals with two booster doses. JN.1 exhibits even greater escape with a 141-fold reduction in neutralization efficiency relative to the ancestral strain. Post-vaccination with an XBB.1.5-adapted mRNA vaccine, neutralization titers against these variants improved but remained significantly lower than those against earlier variants, with JN.1 showing a 27-fold reduction compared to the ancestral strain132, 133.
Covaxin
Bharat Biotech, in collaboration with the National Institute of Virology, developed India’s first indigenous vaccine called Covaxin, which was approved on January 2, 2021, as trials conducted for the vaccine showed promising results with no major adverse effects134. Recent findings suggest effective neutralization of the B.1.1.7 SARS-CoV-2 variant. Interim analysis showed an efficacy of 80.6% for the vaccine135. It is an inactivated vaccine composed of completely inactive SARS-CoV-2 viral particles (RNA surrounded by a protein coat), transformed to prevent replication. It is one of the cheapest COVID-19 vaccines, recommended as a 2-dose regimen with a gap of 28 days between the two shots136. Studies report an increase in virus-specific IgG and other neutralizing antibodies, with a decreased rate of viral replication, after completion of the 2-dose regimen111. Covaxin is stable at 2–8°C and can thus be stored under ordinary refrigeration.
Moderna
The mRNA-1273 vaccine by Moderna is the second vaccine approved by the Food and Drug Administration (FDA) after the BNT162 vaccine by Pfizer and BioNTech. Like BNT162, this is a modified mRNA vaccine encoding a prefusion-stabilized S protein from SARS-CoV-2, which is its major target. The vaccine works by triggering antibody responses, including CD4 T-cell and CD8 cytotoxic T-cell reactions against the viral particle137. Interim analysis conducted after Phase 3 trials reported an efficacy of about 94.5% with no major adverse conditions. However, pain at the injection site, fatigue, and headache or other mild adversities may be observed after vaccination, as in the case of older adults134. The vaccination process follows a two-dose regimen with a gap of 28 days between doses, and unlike the BNT162 vaccine by Pfizer, Moderna’s vaccine can be stored at about -20°C138. As per the reports, the vaccine is best suited for people 18 years or older, providing immunogenicity that lasts for at least 119 days from the first dose139. The Moderna vaccine also produced significant results for pregnant individuals, as a study suggested an equal probability of adverse pregnancies and neonatal outcomes in vaccinated people and those not vaccinated, although consultation regarding the vaccine is a must for pregnant women140.
In response to the evolving viral landscape, updated mRNA vaccines targeting the XBB.1.5 lineage have been developed, and they also demonstrated an increase in neutralizing antibodies against BA.2.86 post-vaccination141. Similarly, Pfizer has also updated its vaccine, showing enhanced immune responses against JN.1 subvariants, including KP.2 and KP.3142. These findings suggest that while the updated boosters improve protection, the degree of efficacy varies among different strains.
Vaccine by Pfizer and BioNTech
BNT162 is the first COVID-19 vaccine approved by the US FDA showing an efficacy of 95% as determined after Phase 3 clinical trial results134. The occurrence of major adverse events was found to be lower in comparison with Moderna’s vaccine, and thus, the Pfizer vaccine was approved for people 16 years and above. It also provides immunogenicity for at least 119 days after the first dose; however, it is much more temperature sensitive than the Moderna vaccine and is preferably stored at -75°C139. The vaccine relies on nucleoside-modified RNA that works against the S protein of the SARS-CoV-2 virus, which is its major virulence factor. This immunization prompts the body to make an antibody response to neutralize the infection, which is reliant upon the S protein for passage through the ACE2 receptors on type 2 alveolar cells. As of 16 May 2021, 85 countries had approved the Pfizer vaccine137.
AZD1222 by AstraZeneca and University of Oxford
AZD1222 COVID-19 vaccine developed by the Oxford University and the British-Swedish pharmaceutical company AstraZeneca is an adenoviral vaccine previously called ChAdOx1. It is a non-replicating chimpanzee adenoviral vaccine, which is also manufactured under the name Covishield in India by the Serum Institute of India using the same formulations137. It was approved by the UK on Dec, 30, 2020, and by India on Jan. 2, 2021. The vaccine works by inducing humoral and cellular responses against the SARS-CoV-2 virus and can be easily stored at 2–8°C134. As per an interim analysis, the vaccine showed an efficacy of about 70.4% after completion of its two-dose regimen with a gap of about 4–12 weeks and was thus recommended for emergency usage in the UK for people 18 years or older.
Management Strategies for Long COVID
There is no single cure for long COVID, and treatment is symptomatic and multidisciplinary. Lifestyle and holistic approaches have been proven beneficial. A balanced diet, adaptively graded exercise, meditation, proper sleep, and consumption of probiotics may assist in ameliorating the symptoms of long COVID. Low doses of naltrexone show a reduction in inflammation and neurological conditions. Serotonin and serotonin-norepinephrine reuptake inhibitors may also help in some cases. Anticoagulants can aid in dissolving microclots, while studies with Remdesivir, Paxlovid, corticosteroids, and monoclonal antibodies are ongoing for long COVID143.
Further, some of the therapeutic developments are underway, such as the RECOVER-VITAL trial investigating whether extended courses of Paxlovid (nirmatrelvir, ritonavir)—an antiviral approved for acute COVID-19—can alleviate long COVID symptoms. Preliminary data from a study involving 13 long COVID patients showed mixed outcomes (9 showed some improvement, 5 reported lasting effects, while 4 saw no improvement). The REVERSE-lc and ADDRESS-LC trials are testing immunomodulatory drugs like baricitinib and bezisterim, aiming to target the immune system rather than the virus. These trials will address the complex immune dysfunction observed in long COVID patients. Meanwhile, RECOVER-NEURO is exploring brain training and stimulation interventions, such as web-based training and a home-based device for transcranial direct stimulation, to address cognitive dysfunction associated with long COVID.
Discussion
The primary focus of this investigation was to assess the impact of COVID-19 on various organ dysfunctions, associated comorbidities, and recent diagnostic advancements. Numerous cases and studies have demonstrated the organ-specific effects of COVID-19, attributed to the presence of ACE2 receptors, leading to ageusia, anosmia, malaise, fatigue, myalgia, and ataxia. Additionally, the virus has been shown to affect the nervous system, causing altered consciousness, seizures, cerebrovascular disease, nerve pain, and vision impairment. Reports have also indicated that both cardiac and renal dysfunction are associated with increased mortality rates.
An elevated expression of ACE2, particularly in bronchial and alveolar cells, among diabetic patients has been linked to enhanced susceptibility to COVID-19 and increased disease severity. Furthermore, immunosuppressive conditions such as cancer, HIV, and pregnancy have been associated with heightened disease severity and mortality. Advanced age, male sex, lower socioeconomic status, and underlying health conditions also serve as major determinants of disease severity. Nevertheless, IC and AA, which act as immune modulators in asthmatic individuals, have been reported to reduce COVID-19 infections in this population.
In the face of rising COVID-19 cases, numerous diagnostic techniques were employed, with RT-PCR serving as the primary detection method. However, interpretation of results requires caution, particularly when testing variants of concern (VOC) such as Alpha and Omicron sublineages BA.4 and BA.5, to avoid false negatives resulting from primer-probe mismatches with the Omicron genome.
Detection of RdRp/HeI, N, and S proteins in Alpha or Omicron cases may yield false negatives for certain sublineages. Therefore, identifying these sublineages can aid in developing a comprehensive diagnostic approach with improved sensitivity99. The S gene target failure (SGTF) has emerged as a key indicator for detecting such cases through next-generation sequencing (Specificity: 98.6–99.8%, Sensitivity: 99.6%)100. With ongoing advancements, additional techniques have been leveraged, leading to the creation of user-friendly kits grounded in the FELUDA and SHERLOCK principles.
Global vaccination campaigns have proven highly effective in reducing the severity of COVID-19 across diverse populations. Moreover, novel vaccine development continues to enhance efficacy and immunogenicity. Data from various studies indicate that declines in antibody titers at 4–6 months are independent of patient-related risk factors, supporting the use of booster doses to bolster immune responses, particularly against emerging strains. Mutations within the SARS-CoV-2 genome can alter the neutralization capacity of vaccine-induced antibodies, thereby influencing vaccine effectiveness. Updated booster formulations increase immune activity against evolving variants such as BA.2.86 and JN.1; however, levels of protection vary, highlighting the urgent need for continued surveillance and vaccine optimization in response to viral evolution133, 144.
Addressing health disparities, expanding healthcare access, and advocating for vaccination among vulnerable groups are vital to mitigating COVID-19's impact globally. In addition, monitoring wastewater for circulating SARS-CoV-2 variants remains imperative. Pilapil .101 reported co-circulation of strains in wastewater, supporting viral mutations and adaptation for transmission and survival. Consequently, surveillance of the viral genome in wastewater serves as a crucial supplement to clinical monitoring.
Conclusions
The elimination of the serious health threat that SARS-CoV-2 poses to humans remains the top priority. In this context, numerous clinical and scientific studies have significantly improved our understanding of genomic architecture, structures, and mutations. This review provides insights into the recent developments regarding disease status in various contexts, such as comorbid conditions and diagnosis, which can assist in revolutionizing medicine and therapeutics for infectious COVID-19. This study highlights the importance of various detection techniques and vaccines for the management of COVID-19 and its comorbidities. Although the WHO declared COVID-19 no longer a global emergency on May 5, 2023, it remains a global health concern requiring ongoing monitoring and booster doses, especially in comorbid cases and with emerging mutant strains. Various treatment options have been shown to be successful in different situations, but further research may help prevent future outbreaks of pandemic-prone strains.
Abbreviations
AA: African American, Ab: Antibody, ACE2: Angiotensin-Converting Enzyme 2, Ag: Antigen, AI: Artificial Intelligence, AXL: AXL Receptor Tyrosine Kinase, BA.1, BA.2, BA.3, BA.4, BA.5: Omicron sub-lineages, B.1.1.7, B.1.351, B.1.427, B.1.429, P.1: SARS-CoV-2 Variants of Concern, BF.7: Omicron sub-lineage (BA.2.75.2), BQ.1, BQ.11: Omicron sub-lineages, CD147: Cluster of Differentiation 147CI: Confidence Interval, CoVs: Coronaviruses, COVID-19: Coronavirus Disease 2019, CRISPR: Clustered Regularly Interspaced Short Palindromic Repeats, CSF: Cerebrospinal Fluid, CT: Computed Tomography, CXR: Chest X-Ray, DC-SIGN: Dendritic Cell-Specific Intercellular adhesion molecule-3-Grabbing Non-integrin, dPCR: Digital Polymerase Chain Reaction, E: Envelope (protein), ELISA: Enzyme-Linked Immunosorbent Assay, FDA: Food and Drug Administration, FELUDA: FnCas9 Editor Linked Uniform Detection Assay, FL: Fusion Loop, GGOs: Ground-Glass Opacities, GRP78: Glucose-Regulated Protein 78, hCoV: Human Coronavirus, HE: Hemagglutinin Esterase, Hel: Helicase, HIV: Human Immunodeficiency Virus, HR1, HR2: Heptapeptide Repeat 1, Heptapeptide Repeat 2, HRT18: Human Rectum Tumor 18 (cell line), IC: Immune Complexes (contextually inferred for asthma treatment), IFN: Interferon, IgG: Immunoglobulin G, LAMP: Loop-Mediated Isothermal Amplification, L-SIGN: Liver/lymph node-Specific ICAM-3 Grabbing Non-integrinM: Membrane (protein), MDA5: Melanoma Differentiation-Associated protein 5, MEGA: Molecular Evolutionary Genetics Analysis, MERS-CoV: Middle East Respiratory Syndrome Coronavirus, MIS-C: Multisystem Inflammatory Syndrome in Children, ML: Maximum Likelihood, Mpro: Main Protease (3CLpro), mRNA: Messenger RNA, N: Nucleocapsid (protein), NASBA: Nucleic Acid Sequence-Based Amplification, NCBI: National Center for Biotechnology Information, NiRAN: Nidovirus RdRp-Associated Nucleotidyltransferase, NSPs: Non-Structural Proteins, NTD: N-Terminal Domain, ORF: Open Reading Frame, ORF1a, ORF1b: Open Reading Frame 1a, 1b, PAM: Protospacer Adjacent Motif, PCR: Polymerase Chain Reaction, PLpro: Papain-Like Protease, POC: Point-Of-Care, pp1a, pp1ab: Polyprotein 1a, Polyprotein 1ab, RAAS: Renin-Angiotensin-Aldosterone System, RAS: Renin-Angiotensin System, RBD: Receptor-Binding Domain, RdRp: RNA-dependent RNA Polymerase, RIG-I: Retinoic acid-Inducible Gene-I, RNA: Ribonucleic Acid, R₀: Basic Reproduction Number, RT-PCR: Reverse Transcription Polymerase Chain Reaction, RTC: Replication-Transcription Complex, S: Spike (protein), S1, S2: Spike subunit 1, Spike subunit 2, SARS-CoV: Severe Acute Respiratory Syndrome Coronavirus, SARS-CoV-2: Severe Acute Respiratory Syndrome Coronavirus 2, SD1, SD2: Subdomain 1, Subdomain 2, SGTF: S Gene Target Failure, sgmRNA: Sub-genomic messenger RNA, SHERLOCK: Specific High-sensitivity Enzymatic Reporter unLOCKing, SNPs: Single Nucleotide Polymorphisms, ssRNA: Single-Stranded RNA, STAT1/2: Signal Transducer and Activator of Transcription 1/2, TANK: TRAF Family Member-associated NF-kappa-B activator, TBK1/IKKε: TANK-Binding Kinase 1 / Inhibitor of nuclear factor kappa-B kinase subunit epsilon, TIMP1: TIM-1 (T-cell Immunoglobulin and Mucin domain-containing protein 1), TMPRSS2: Transmembrane Serine Protease 2, TM: Transmembrane (domain), TRAF3: TNF Receptor-Associated Factor 3, UTR: Untranslated Region, VOC: Variant of Concern, WHO: World Health Organization, XBB: Omicron recombinant lineage
Acknowledgments
None.
Author’s contributions
Methodology, JG., PV., SS, NS and AY.
Funding
None.
Availability of data and materials
Data and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Declaration of generative AI and AI-assisted technologies in the writing process
The authors declare that they have not used generative AI (a type of artificial intelligence technology that can produce various types of content including text, imagery, audio and synthetic data. Examples include ChatGPT, NovelAI, Jasper AI, Rytr AI, DALL-E, etc) and AI-assisted technologies in the writing process before submission.
Competing interests
The authors declare that they have no competing interests.

