Transcriptome Analysis of the Protective Mechanism of Ectoine on Human Skin Keratinocytes
- Department of Basic Medical Sciences, Medical College, Qinghai University, Xining 810016, China
- College of Life Sciences, Northwest A&F University, Yangling, Shaanxi 712100, China
- School of Civil Engineering and Water Resources, Qinghai University, Xining, 810016, China
Abstract
Introduction: Ectoine, a natural protective agent produced by halophilic bacteria, demonstrates remarkable skin-protective efficacy; however, the precise molecular mechanisms underlying its effects on skin cells remain poorly understood. This study investigated the differentially expressed genes (DEGs) in human keratinocytes (HaCaT) treated with ectoine to clarify its protective mechanisms.
Methods: Cell viability was evaluated using the CCK-8 assay, identifying 0.2 mg/mL as the optimal ectoine concentration. A control group (C, 0 mg/mL) and an ectoine group (E, 0.2 mg/mL) were established. Flow cytometry quantified reactive oxygen species (ROS) and apoptosis. Illumina HiSeq RNA-seq identified DEGs; selected genes were validated by RT-qPCR.
Results: Ectoine at 0.2 mg/mL increased cell viability to 134.0 % ± 3.2. Compared with the control group, ectoine significantly reduced the apoptosis rate and intracellular ROS levels (P ≤ 0.05). RNA-seq (n = 3) identified 292 DEGs (87 up-regulated, 205 down-regulated). Among them, MMP25, NOXO1, ANGPTL4, and FoXO6—genes involved in oxidative stress, apoptosis, and proliferation—were markedly down-regulated, suggesting enhanced proliferation and anti-oxidative, anti-apoptotic effects. The cell-adhesion gene CNFN was significantly up-regulated, potentially reducing mechanical damage.
Conclusion: Ectoine serves as a potent stabilizer and protectant, safeguarding skin by modulating genes that regulate oxidative stress, proliferation, and apoptosis.
Introduction
Human keratinocyte cells (HaCaT), the primary cells of the skin epidermis, play a protective role by forming epidermal structures. When external factors such as dryness, radiation, wind, and temperature fluctuations accumulate, HaCaT cells are the first to be compromised, leading to skin dryness and aging-related damage1.
Ectoine, a natural protectant synthesized by halophilic bacteria, shields these organisms from high-osmolarity environments2. It stabilizes cell membranes, nucleic acids, and proteins and lessens cellular stress caused by external factors3, 4, 5, 6, making it superior to other compatible solutes7. Recent studies show that ectoine binds water effectively and prevents transepidermal water loss8. For instance, Hseu .9 reported that ectoine promotes skin whitening by inhibiting the ROS-p53/POMC-α-MSH pathway in UV-irradiated HaCaT cells. Cheng .10 used UVA- and HO-induced oxidation models of human skin fibroblasts and found that ectoine upregulated PI3K/AKT-related genes (COL1A1, COL1A2, FN1, IGF2, NR4A1, PIK3R1) and reduced intracellular ROS and malondialdehyde (MDA), thereby exerting protective effects. Moreover, ectoine alleviates atopic dermatitis and retinoid dermatitis caused by a weakened skin barrier11. However, the global genetic changes induced by ectoine in HaCaT cells remain unclear.
With the widespread adoption of high-throughput sequencing, transcriptomics has become a powerful tool for exploring quorum sensing, phenotypic shifts, metabolic adaptation, and key gene regulation. Lin .12 showed that vitamin D markedly protects against skin photoaging; single-cell sequencing revealed that vitamin D downregulated inflammatory genes (Cd74, Cxcl2, Pcolce, Procr). RNA-seq data also demonstrated that ultraviolet A (UVA) activates Nrf2 and its targets—cyclin D1, TNFrsf1b, and Mybl1—producing antioxidant, anti-inflammatory, and anticancer effects in UVB-induced non-melanoma skin cancer (NMSC).
Therefore, we investigated the effects of ectoine on HaCaT cell viability, apoptosis, and intracellular ROS. We then applied transcriptomics to identify ectoine-induced genetic alterations and validated key findings with RT-qPCR. Our results show that ectoine promotes proliferation, inhibits apoptosis, and scavenges free radicals in HaCaT cells, providing mechanistic insight and supporting its therapeutic application.
Methods
Reagents and instruments
HaCaT cells were purchased from Wuxi Xinrun Biotechnology Co., Ltd. (Wuxi, China). Ectoine (≥98 %) was purchased from Shanghai Titan Scientific Co., Ltd. (Shanghai, China). Dulbecco’s modified Eagle’s medium (DMEM), TRIzol reagent, PrimeScript™ RT reagent kit with gDNA Eraser, and TB Green® Premix Ex Taq™ kit were obtained from TAKARA Holdings Inc. (Japan). Cell Counting Kit-8 was purchased from Elabscience Biotechnology Co., Ltd. (Wuhan, China). Annexin V-FITC Apoptosis Detection kit was purchased from TransGen Biotech Co., Ltd. (Beijing, China). The reactive oxygen species (ROS) test kit was purchased from Wuhan Servicebio Technology Co., Ltd. (Wuhan, China). An xMark microplate reader was bought from Bio-Rad Laboratories (USA). A CO incubator (BPN-150CH) was purchased from Thermo Fisher Scientific (USA); a CytoFLEX flow cytometer was obtained from Beckman Coulter, Inc. (USA); and a LightCycler® 96 real-time PCR system was purchased from F. Hoffmann-La Roche Ltd. (Switzerland). Qubit 4.0 and NanoDrop 2000 spectrophotometers were manufactured by Thermo Fisher Scientific (USA). An Agilent 5300 Bioanalyzer was provided by Agilent Technologies Inc. (USA). A NovaSeq X Plus sequencer was purchased from Illumina, Inc. (USA).
Cell culture
HaCaT cells were cultured in high-glucose DMEM supplemented with 10 % fetal bovine serum (FBS) and 1 % penicillin–streptomycin at 37 °C in 5 % CO. Cells were observed under an inverted microscope, and experiments were performed when confluence reached 90–95 %.
Filtering the optimum concentration of ectoine
HaCaT cells were seeded into 96-well plates (5 × 10³ cells well⁻¹) and incubated for 24 h at 37 °C, 5 % CO. After washing with PBS, cells were treated with a concentration gradient of ectoine (0.05–0.50 mg mL⁻¹; n = 5 per group) for 24 h. Cell viability was assessed with the CCK-8 assay to determine the optimal ectoine concentration.
Detecting apoptosis rate and intracellular ROS
HaCaT cells were seeded in 6-well Lab-Tek chambers with complete DMEM and grown to 80 % confluence. Cells were divided into a control group (C, 0 mg mL⁻¹) and an ectoine group (E, 0.20 mg mL⁻¹) (n = 3 per group). Cells were centrifuged at 1000 rpm for 5 min and washed with PBS. One aliquot was resuspended in 100 µL Annexin V Binding Buffer, supplemented with 5 µL Annexin V-FITC and 5 µL PI, and incubated for 15 min at room temperature in the dark. The remaining cells were incubated with 10 µmol L⁻¹ DCFH-DA in serum-free medium at 37 °C for 30 min in the dark. After two PBS washes, cells were filtered through a 300-mesh nylon membrane and analyzed on a flow cytometer to measure apoptosis and ROS levels.
Library construction and quality control of transcriptome
Total RNA from groups C and E (n = 3 per group) was extracted using TRIzol, and integrity was verified on 1 % agarose gels. RNA purity (OD = 1.8–2.2, OD ≥ 2.0) and integrity (RIN ≥ 6.5, 28S:18S ≥ 1.0; >1 µg) were confirmed on a NanoDrop 2000 and Agilent 5300, respectively. Double-stranded cDNA was synthesized with the SuperScript kit and random hexamers, size-selected to ≈300 bp, and amplified by PCR. Strand-specific libraries were prepared, validated, and quantified on a Qubit 4.0 and sequenced on a NovaSeq X Plus. Adapter-containing reads, reads with >50% bases of Q ≤ 20, or containing undetermined bases were removed with fastp (v0.23.4)13. Clean reads were aligned to the reference genome using HISAT2 (v2.2.1)14 and assembled with StringTie15 .
Differential expression analysis and functional enrichment
Transcripts per million reads (TPM) was estimated with RSEM16. Differential expression was analyzed with DESeq2 (v1.46.0)17 or DEGseq (v1.26.0)18; genes with |log₂FC| ≥ 1 and FDR ≤ 0.05 (DESeq2) or FDR ≤ 0.001 (DEGseq) were considered significantly differentially expressed. GO and KEGG enrichment were performed with GOATOOLS and KOBAS, respectively; terms with a Bonferroni-corrected P ≤ 0.05 were deemed significant19.
RT-qPCR validation of key DEGs
RNA samples meeting the above quality criteria (concentration > 5 ng µL⁻¹) were reverse-transcribed using the PrimeScript™ RT kit with gDNA Eraser. The 20 µL reverse-transcription mix contained 10 µL Reaction I (5 × gDNA Eraser Buffer 2 µL, gDNA Eraser 1 µL, RNA + RNase-free H₂O to 7 µL); incubation was 42 °C for 2 min. Reaction II (PrimeScript RT Enzyme Mix I 1 µL, RT Primer Mix 1 µL, 5 × PrimeScript Buffer 2 4 µL, RNase-free H₂O 4 µL) was then added. cDNA was diluted 1:20 for qPCR. The 20 µL qPCR mix contained 10 µL TB Green Premix Ex Taq II, 0.8 µL each primer (10 µM), 2 µL cDNA, and 6.4 µL H₂O. Cycling: 95 °C 3 min; 45 cycles of 95 °C 10 s, 58 °C 20 s, 72 °C 30 s, with fluorescence acquisition during extension. PPIA served as the reference gene (n = 3 per group). Relative expression was calculated by the 2 method. Primers were synthesized by Sangon Biotech (
Primer sequences and product size
|
Genes name |
Primer sequences (5'→3') |
Length (bp) | |
|
ANGPTL4 |
F: GAGGCTGGACAGTAATTCAGA, R:CGTGATGCTATGCACCTTCT |
134 | |
|
MMP25 |
F: GACTGGCTGACTCGCTATGG, R:TGATGGCATCGCGCAACTT |
88 | |
|
NOXO1 |
F: ATTCAGGCAGCTCAAGACCC, R:TGACCGAGAAGCTTTGGGAG |
93 | |
|
CNFN |
F: TTGCTCCTCTGTGCCTTGCC, R:ACGGAGCCCTGGATGTGGT |
130 | |
|
FoxO6 |
F: GTGGGGGAACCTTTCCTACG, R:TTCTGCACGCGGATGAACC |
207 | |
|
PPIA |
F: GTCAACCCCACCGTGTTCTT, R:GTCAACCCCACCGTGTTCTT |
97 | |
Statistics
Statistical analyses were performed in GraphPad Prism 8. Normality was tested by the D'Agostino–Pearson omnibus test. Two-tailed Student’s t-tests compared two groups, and one- or two-way ANOVA was used for ≥3 groups. Data are reported as mean ± SD (ns, not significant; * < 0.05; ** < 0.01; *** < 0.001; **** < 0.0001).
Results
Ectoine promoted the proliferation of HaCaT cells
Cell viability was assessed after treating HaCaT cells with different concentrations of ectoine. Results showed that cell viability increased as ectoine concentrations rose from 0.05 mg/mL to 0.20 mg/mL (103.94 ± 1.95 and 134.00 ± 3.22, respectively), and then declined as the concentration increased from 0.20 mg/mL to 0.50 mg/mL (107.10 ± 2.47; Figure 1). Overall, cell viability was significantly increased (>100 %) in all ectoine-treated groups (Table S1), indicating that ectoine may enhance the proliferation of HaCaT cells. Consequently, based on experimental results, 0.20 mg/mL was determined to be the optimal concentration for further experiments.
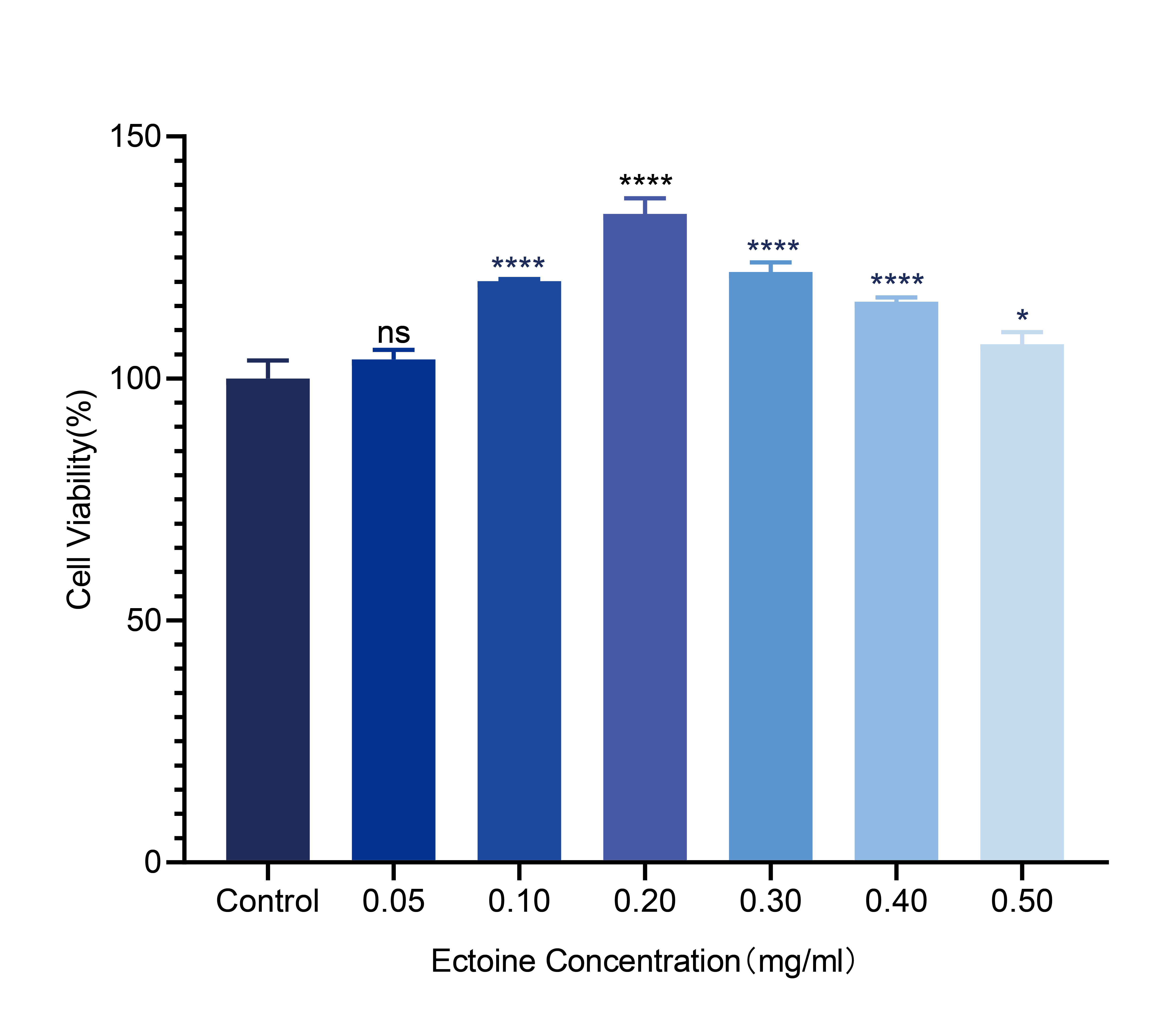
Effect of different concentrations of ectoine on the viability of HaCaT cells. Error bars indicate the standard deviation of three parallel samples. *:
Ectoine inhibited apoptosis and ROS generation in HaCaT cells
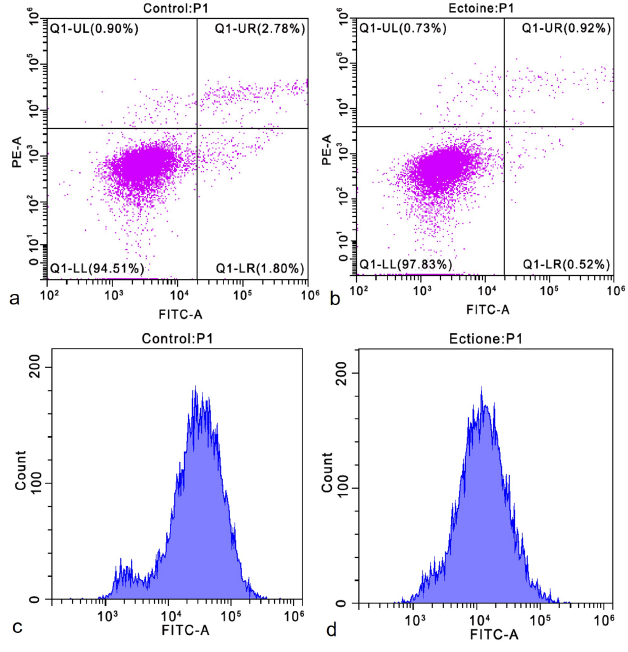
Apoptosis rates were analyzed using flow cytometry (Figure 2ab) after treating cells with 0.20 mg/mL ectoine. The apoptosis rate in the control group was 4.65 ± 0.57 %, whereas it was significantly lower in the ectoine-treated group (1.91 ± 0.19 %; Table S2). This suggests that ectoine may regulate HaCaT cell apoptosis by exerting anti-apoptotic effects. Intracellular reactive oxygen species (ROS) levels were measured using flow cytometry (Figure 2cd). The relative DCFH-DA fluorescence intensity was significantly decreased in the ectoine-treated group, indicating a substantial reduction in intracellular ROS levels (Table S3). These findings demonstrate that ectoine effectively reduces endogenous ROS and minimizes oxidative stress.
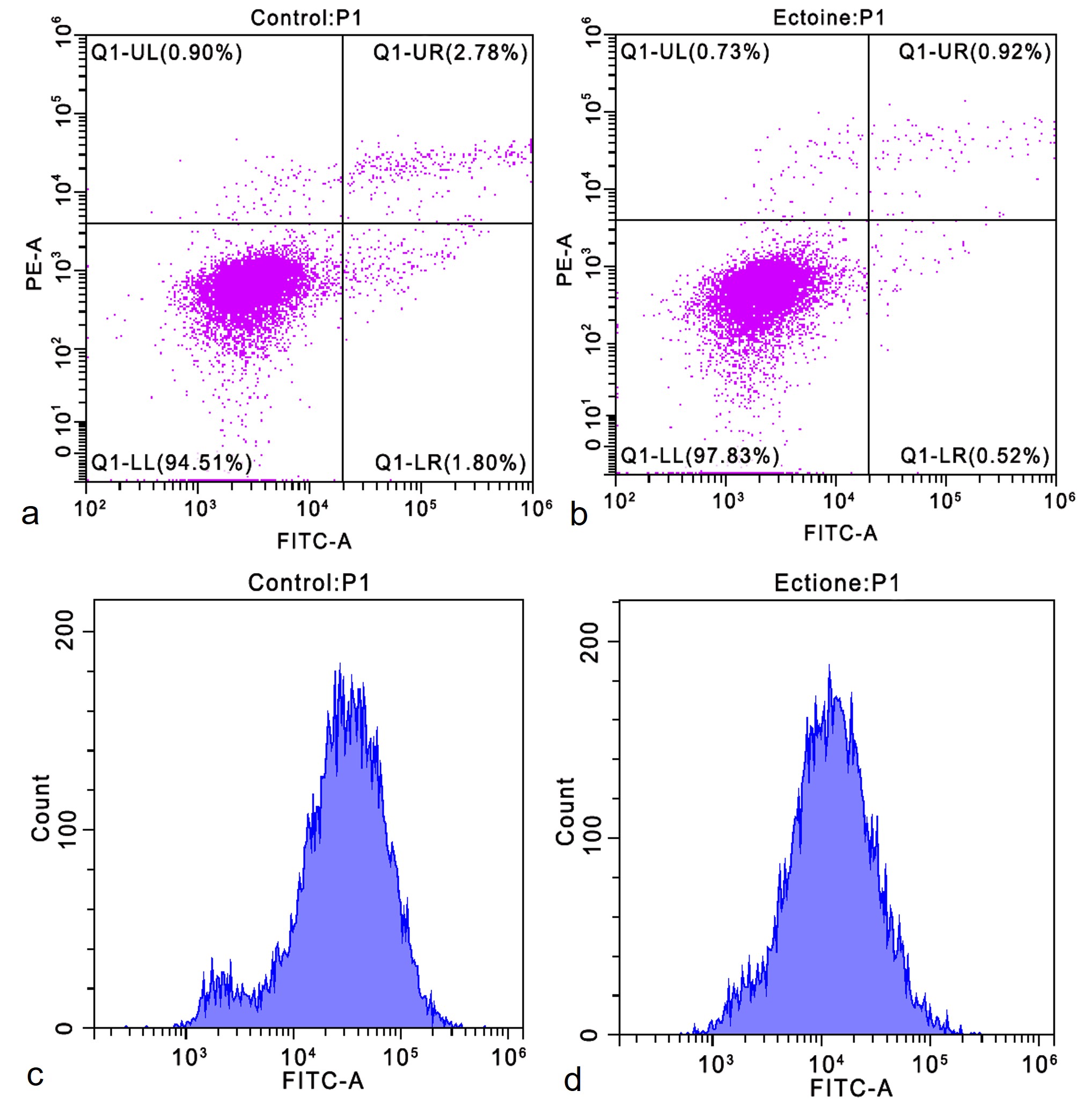
Apoptosis rate and intracellular ROS level of HaCaT cells. Apoptosis rate of cells treated with 0 mg/ml ectoine for 24 h (a). The apoptosis rate of cells treated with 0.20 mg/ml ectoine for 24 h (b). The intracellular ROS level in cells treated with 0 mg/ml ectoine for 24 h (c). The intracellular ROS level in cells treated with 0.20 mg/ml ectoine for 24 h (d). Abbreviations: HaCaT (Human Keratinocytes Cells), ROS (Reactive Oxygen Species)
Transcriptome data processing and quality-control analysis
The raw data were filtered and screened to obtain more than 41,204,442 clean reads per group, with Q30 > 93.99 % and Q20 > 97.90 %. The error rate was <0.03 %, and the GC content ranged from 51.22 % to 52.79 % (
Filtered data and quality statistics
|
Sample |
Raw reads |
Clean reads |
Q20 (%) |
Q30 (%) |
GC (%) |
Total mapped |
Multiple mapped |
Unique mapped |
|
C1 |
47683022 |
47205576 |
98.02 |
94.29 |
52.36 |
97.48% |
3.03% |
94.45% |
|
C2 |
43301474 |
42862366 |
97.9 |
93.99 |
51.9 |
97.39% |
3.07% |
94.32% |
|
C3 |
53243942 |
52725454 |
97.97 |
94.17 |
52.79 |
97.55% |
3.33% |
94.22% |
|
E1 |
41565756 |
41204442 |
97.99 |
94.21 |
51.05 |
97.53% |
3.47% |
94.06% |
|
E2 |
46817134 |
46392670 |
98.05 |
94.4 |
51.49 |
97.52% |
3.44% |
94.09% |
|
E3 |
52862954 |
52385586 |
98.08 |
94.45 |
51.22 |
97.50% |
4.10% |
93.40% |
Analysis of gene expression in response to ectoine
The expression levels of DEGs in the control (C) and ectoine (E) groups, measured as log2(FPKM + 1), were subjected to hierarchical clustering. Ectoine reversed the transcriptional differences observed between the C and E groups: clusters with low expression in the C group shifted to high expression in the E group, whereas clusters with high expression in the C group became low in the E group. More low-expression than high-expression clusters were observed in the E group, suggesting that ectoine primarily suppresses overall gene expression in HaCaT cells (Figure 3a). Differential-expression analysis (log2 fold-change ≠ 0, P ≤ 0.05) identified 292 DEGs between C and E, including 87 up-regulated and 205 down-regulated genes (Figure 3b).
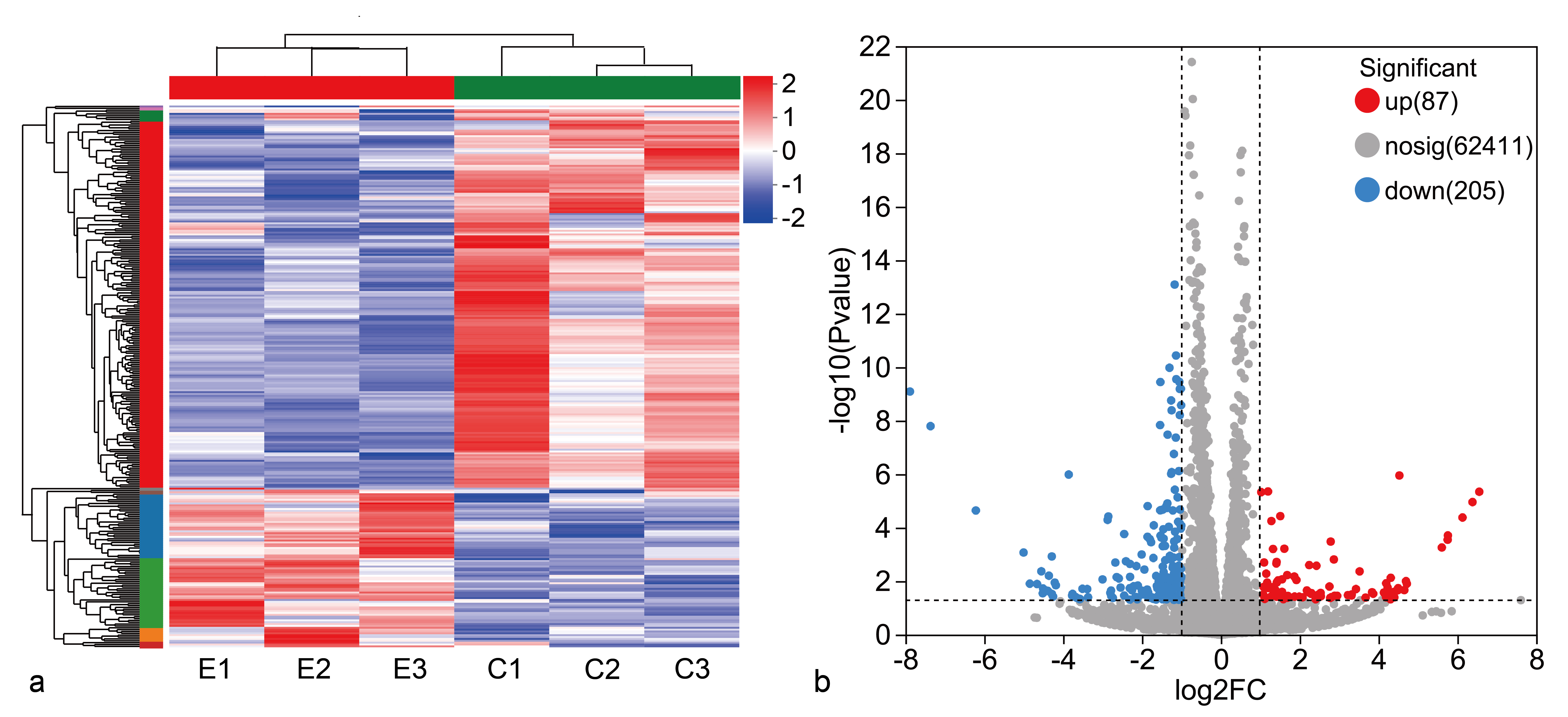
Statistical analysis of differential gene data in the comparison group. Cluster analysis of DEGs in different groups (a). Volcano plot of differential genes (b). Abbreviation: DEGs (differential expression genes)
GO and KEGG functional annotation analysis of differential genes
DEGs between the C and E groups were annotated using the Gene Ontology (GO) database. In total, 292 DEGs were enriched in 43 GO terms. The top 20 enriched terms fell into three categories—biological process (BP), cellular component (CC), and molecular function (MF; Figure 4a). Within BPs, DEGs were mainly enriched in cellular processes, biological regulation, and metabolic processes. For CCs, DEGs were largely enriched in cell parts, organelles, and membrane components. Regarding MFs, DEGs were primarily linked to binding and catalytic activities.
Mapping DEGs to the KEGG pathway database grouped them into five categories (top 20 pathways shown). In Metabolism (21 DEGs), genes were mainly involved in carbohydrate and lipid metabolism. In Environmental Information Processing (32 DEGs), genes were related to signal transduction and signaling-molecule interactions. In Cellular Processes (21 DEGs), genes concerned cell growth, death, and eukaryotic communities. In Organismal Systems (64 DEGs), genes were involved in the immune, endocrine, and nervous systems. In Human Diseases (69 DEGs), genes were mainly associated with cancer and endocrine and metabolic diseases (Figure 4b).
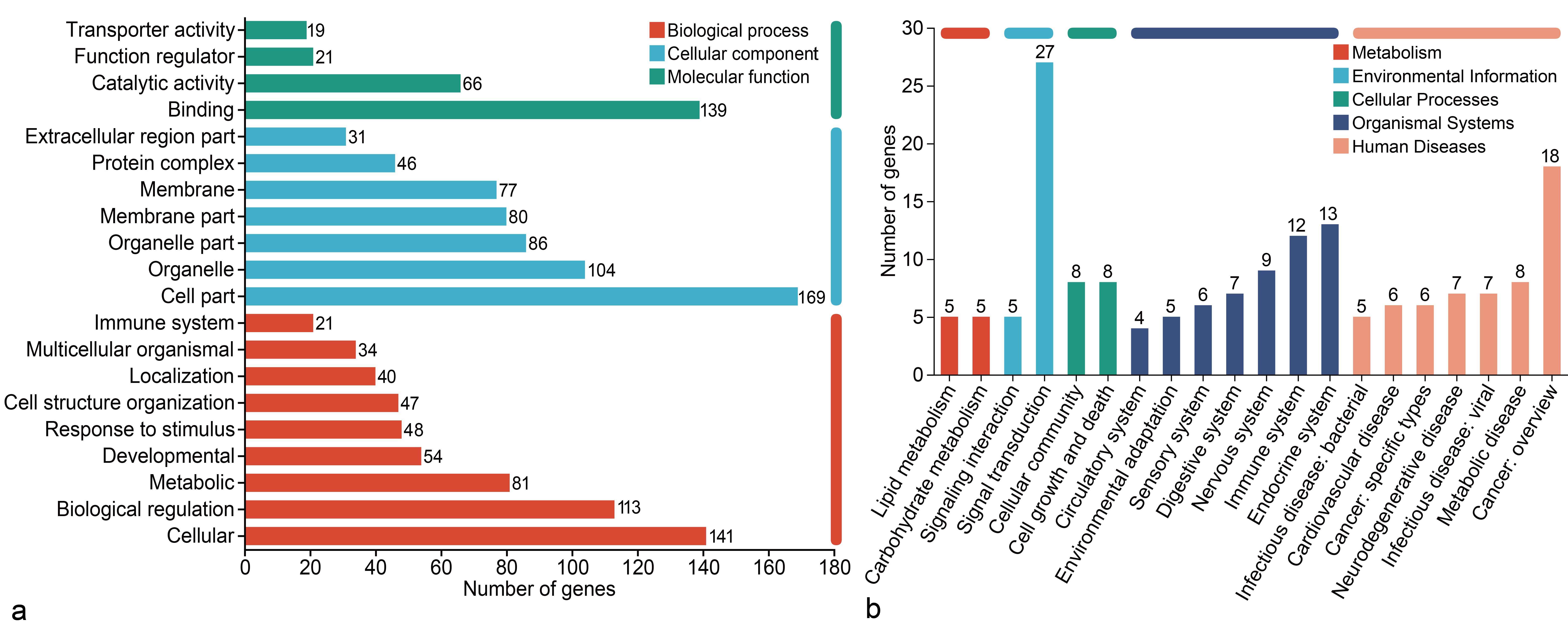
Functional category of DEGs in HaCaT Cells. GO functional annotation analysis (a). KEGG functional annotation analysis (b). Abbreviations: HaCaT (Human Keratinocytes Cells), DEGs (differential expression genes), GO (Gene Ontology), KEGG (Kyoto Encyclopedia of Genes and Genomes)
GO and KEGG functional enrichment analysis of differential genes
GO enrichment identified the 20 most significantly enriched terms, primarily associated with molecular functions (3) and biological processes (17; Figure 5a). Five GO terms showed the greatest enrichment: glutamine catabolic process (rich factor = 0.40; 2 DEGs), glutaminase activity (0.40; 2 DEGs), interleukin-21-mediated signaling pathway (0.333; 2 DEGs), glutamate biosynthetic process (0.333; 2 DEGs), and regulation of oocyte maturation (0.333; 2 DEGs).
KEGG enrichment analysis showed that DEGs participated in 208 signaling pathways. The 20 pathways with the most significant enrichment were analyzed (Figure 5b); the Ras signaling pathway contained the largest number of enriched genes (Figure 5c). The Ras pathway is a crucial intracellular signaling mechanism involved in cell growth, differentiation, apoptosis, and metabolism. Ectoine appears to act by activating receptor tyrosine kinases (RTKs) on the cell surface, triggering downstream cascades that lead to Ras activation. Activated Ras mediates anti-apoptotic effects via the PI3K/Akt pathway: the Ras-PI3K/Akt axis suppresses the AFX transcription factor, thereby down-regulating Fas ligand (FasL) expression, and also promotes NF-κB signaling, collectively regulating apoptosis, cell survival, and growth.
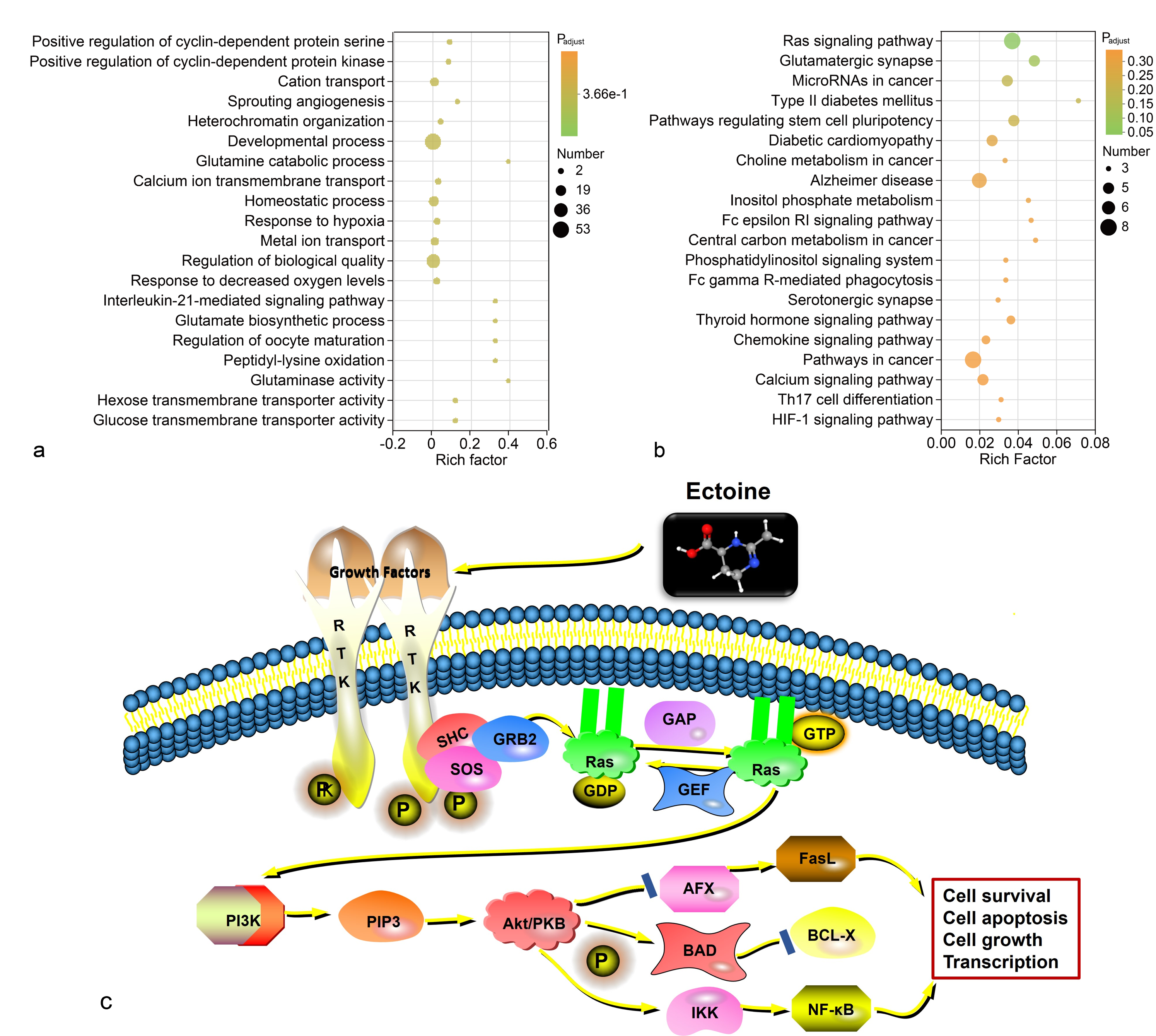
GO and KEGG enrichment analysis of the DEGs. Bubble chart of GO enrichment analysis (a). Bubble chart of KEGG enrichment analysis (b). Analysis diagram of the potential mechanism of ectoine regulating the Ras pathway (c). Abbreviations: DEGs (differential expression genes), DEGs (differential expression genes), GO (Gene Ontology), KEGG (Kyoto Encyclopedia of Genes and Genomes)
Verification of key DEGs related to skin-cell protection using RT-qPCR
Ectoine treatment markedly enhanced HaCaT cell viability while reducing apoptosis and ROS levels. Based on the transcriptomic data, five key DEGs associated with oxidation, apoptosis, and proliferation were selected for RT-qPCR validation (Table S4): MMP25 (extracellular-matrix degradation), NOXO1 (endogenous ROS generation), ANGPTL4 (inflammatory oxidative stress), FOXO6 (pro-apoptotic), and CNFN (keratinocyte structural protein). RT-qPCR showed that ANGPTL4 expression was significantly down-regulated (Figure 6a), with lower levels than indicated by RNA-seq. MMP25, NOXO1, and FOXO6 were also significantly down-regulated (Figure 6b–d), whereas CNFN was significantly up-regulated (Figure 6e) in ectoine-treated cells, consistent with RNA-seq findings. Overall, RT-qPCR results corroborated the RNA-seq data regarding up- and down-regulation patterns (Figure 6f). The accuracy of gene-expression quantification can be affected by primer design, instrument sensitivity, and reagent quality.
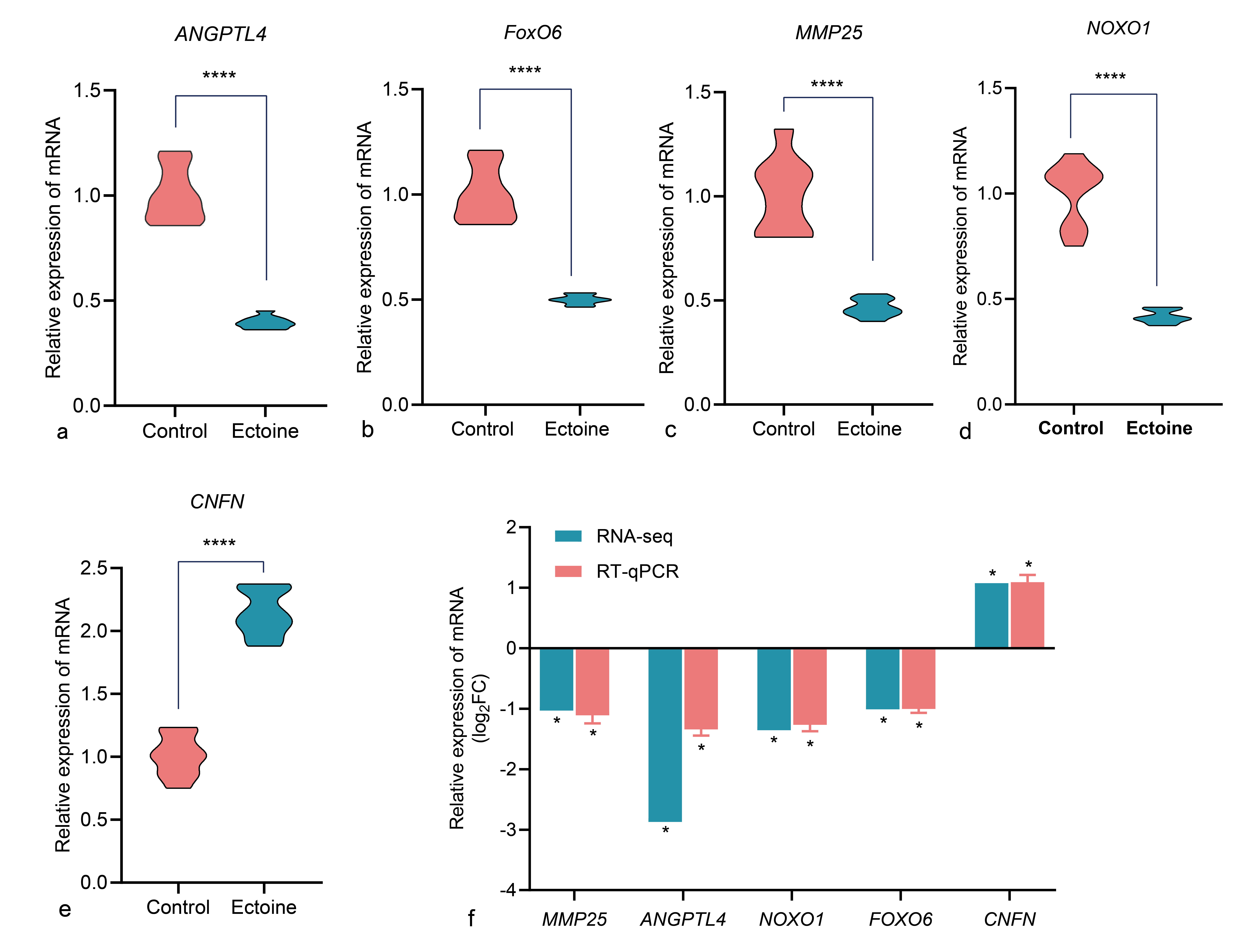
Expression of select genes in Control and ectoine HaCaT cells, data were presented as the mean ± standard error (n = 3/group). Comparing the differences in ANGPTL4, FoxO6, MMP25, NOXO1, and CNFN mRNA expression between the cells in each group, the ectoine treatment group was compared with the control group (a-e). Compare the results of RT-qPCR with those of RNA-Seq (f). Abbreviation: ANGPTL4 (Angiopoietin-like 4), FoxO6 (Forkhead box O6), MMP25 (Matrix metallopeptidase 25), NOXO1 (NADPH oxidase organizer 1), and CNFN (Cornifelin), mRNA (Messenger RNA), RT-qPCR (Quantitative reverse transcription polymerase chain reaction), RNA-Seq (RNA sequencing)
Discussion
Ectoine protects the skin by reducing the apoptosis rate and ROS of HaCaT cells
Apoptosis can be induced by altering the external environment, such as intense UV irradiation and desiccation, by generating ROS. ROS-induced cellular oxidative stress can cause mitochondrial damage, leading to calcium-ion imbalance and inducing the release of apoptotic proteins from the cells, which initiates the process of apoptosis and accelerates the process of skin aging20, 21. Hseu .9 found that after pretreatment of HaCaT cells with ectoine, the expression of antioxidant proteins (HO-1, NQO-1, γ-GCLC) and the catalytic subunit of γ-glutamylcysteine ligase (γ-GCLC) was up-regulated, thereby inhibiting the production of ROS. Cheng 10 observed that ectoine alleviated oxidative damage in human skin fibroblasts by up-regulating genes associated with the PI3K/AKT signaling pathway, such as COL1A1, COL1A2, FN1, IGF2, NR4A1 and PIK3R1. In skin cells, NADPH oxidases (NOXs) are endogenous sources of ROS production, while the NOX1 holoenzyme is the main source of ROS produced by ultraviolet (UV) radiation22. NOXO1, an essential subunit of NOX1 (encoded by the gene NOXO1), plays a crucial role in activating NOX1 enzymes. ROS also activates signaling pathways that promote the synthesis of matrix metalloproteinases (MMPs)23, 24. MMP25 is a key gene encoding MMPs, which are enzymes responsible for the degradation of extracellular-matrix (ECM) proteins25. ECM proteins are the main connective-tissue component of the dermis26. Degradation of the ECM leads to skin laxity and reduced elasticity, which accelerates skin aging and impairs barrier function. In the ectoine experimental group of this study, significant down-regulation of the expression of the gene NOXO1 can reduce the expression of the NOX1 holoenzyme, thus reducing the production of ROS, while significant down-regulation of the expression of the gene MMP25 can decrease the rate of ECM degradation, thereby mitigating ROS-induced damage to skin cells and delaying skin aging. Unlike Hseu , who focused on the induction of antioxidant proteins, we identify NOXO1 as a critical target gene of ectoine, demonstrating that its suppression disrupts the assembly of the NOX1 holoenzyme—an effect that directly inhibits ROS generation at an upstream stage rather than scavenging it downstream. While Cheng reported that ectoine up-regulates COL1A1 and COL1A2 through the PI3K/AKT pathway, we uncover an additional mechanism by which ectoine down-regulates MMP25 expression, thereby slowing ECM degradation—a previously unrecognized mechanism that contributes to the preservation of skin structural integrity under oxidative stress.
Research has demonstrated that ectoine maintains the integrity of the corneal barrier by inhibiting the pro-inflammatory response and promoting the expression of cytokine IL-3727. ANGPTL4 belongs to the family of ANGPTL proteins, which are involved in vasculature generation, inflammation, oxidative stress, vascular permeability and wound healing28, 29, 30. In a psoriasis study, the gene ANGPTL4 was found to promote inflammatory responses (TNF-α, IL-1β, IL-6 and IL-17A) in keratinocytes by regulating ERK1/2- and STAT3-dependent signaling pathways31. Conversely, silencing ANGPTL4 inhibits these effects. Forkhead box protein O6 (FoxO6) is a member of the FoxO family of nuclear transcription factors, which play a crucial role in regulating a diverse array of cellular processes. These processes encompass key functions in cell survival, proliferation, apoptosis, metabolic homeostasis and aging regulation32, and their target genes include TRAIL (TNF-related apoptosis-inducing ligand), FasL (Fas ligand), Bim (Bcl-2-interacting cell death mediator), pro-apoptotic Bcl-2 family members and Bcl-633, 34. Importantly, FoxO6 phosphorylation at Ser184, mediated by Akt, has been shown to promote oxidative stress and inflammation through NF-κB activation in keratinocytes35. This finding is consistent with our observation that ectoine exerts anti-apoptotic and antioxidant effects by down-regulating FoxO6. Notably, FoxO6 knockdown has been reported to protect ARPE-19 cells from high-glucose-induced ROS accumulation36 and cardiomyocytes from hypoxia-induced damage37, further supporting its broad regulatory role in cellular stress responses. In this study, the expression of ANGPTL4 was significantly down-regulated in the ectoine experimental group, suggesting that ectoine inhibits the inflammatory response and thus maintains corneal-barrier integrity. Additionally, the expression of FoxO6 was significantly down-regulated, indicating that ectoine reduces the rate of apoptosis by modulating apoptosis-related transcription factors and by reducing oxidative stress and inflammation.
In this study, 0.2 mg ml⁻¹ ectoine significantly reduced the apoptosis rate and ROS levels in HaCaT cells, and, combined with the transcriptomics results, we speculate that ectoine may protect cells and delay aging by reducing the expression of genes associated with ROS production and apoptosis.
Ectoine protects skin by enhancing HaCaT cell adhesion
The epidermis is the outermost protective barrier of the human body, and this important barrier function is largely attributed to the differentiation and maturation of keratinocytes and their intercellular-adhesion properties38. Ectoine can enhance the mobility of the HaCaT-cell membrane by increasing hydrophilicity and molecular spacing, which in turn enhances the repair mechanism of the membrane and stabilizes the skin barrier39, 40. Cornifelin (CNFN) has been identified as a protein component of epidermal corneocytes41. Liu .42 found that CNFN deficiency contributes to cyclic alopecia and impairs the skin’s functional barrier in Zdhhc13skc4 mice. Wagner .43 found that CNFN deficiency results in defects in keratinocyte desmosome formation, reduced intercellular adhesion and increased susceptibility to damage in the epidermal epithelial layer. In this study, transcription-analysis results indicated that CNFN expression was significantly up-regulated in the experimental group, suggesting that ectoine may increase cell adhesion and connectivity by up-regulating CNFN, thereby enhancing the mechanical stability of cells. This represents a novel finding in elucidating ectoine’s skin-repair mechanism.
While this study focused on ectoine’s cytoprotective effects in HaCaT cells, we acknowledge the limitation of using a single cell type and propose future research with protein-level validation of key DEGs, multi-cell systems, additional markers of oxidative stress and tissue models to fully characterize its potential in skin-barrier protection, anti-apoptosis, antioxidant activity and other therapeutic applications.
Conclusions
Ectoine significantly enhances HaCaT cell viability, reduces oxidative stress and apoptosis, and promotes cell proliferation. Transcriptome analysis revealed that ectoine modulates key genes (MMP25, NOXO1, ANGPTL4, FoXO6, and CNFN) and the RAS pathway, enhancing antioxidative and antiapoptotic effects while strengthening cell adhesion. These findings demonstrate that ectoine protects skin by regulating genes involved in the oxidative response, proliferation, and apoptosis.
Abbreviations
Akt: Ak strain transforming, ANGPTL4: Angiopoietin-like 4, ANOVA: Analysis of Variance, BP: Biological Process, C: Control Group, CC: Cellular Component, CCK-8: Cell Counting Kit-8, cDNA: Complementary DNA, CNFN: Cornifelin, COL1A1: Collagen Type I Alpha 1 Chain, COL1A2: Collagen Type I Alpha 2 Chain, Ct: Threshold Cycle, DCFH-DA: 2',7'–Dichlorofluorescin diacetate, DEGs: Differentially Expressed Genes, DESeq2: Differential Gene Expression analysis based on the Negative Binomial distribution, DMEM: Dulbecco's Modified Eagle's Medium, DNA: Deoxyribonucleic Acid, E: Ectoine Group, ECM: Extracellular Matrix, ERK: Extracellular Signal-Regulated Kinase, F: Forward, FasL: Fas Ligand, FBS: Fetal Bovine Serum, FC: Fold Change, FDR: False Discovery Rate, FN1: Fibronectin 1, FoxO6: Forkhead box protein O6, FPKM: Fragments Per Kilobase of transcript per Million mapped reads, gDNA: Genomic DNA, GC: Guanine-Cytosine, GO: Gene Ontology, γ-GCLC: Gamma-Glutamyl-Cysteine Ligase Catalytic Subunit, HaCaT: Human adult Keratinocyte cell line, IGF2: Insulin Like Growth Factor 2, IL: Interleukin, KEGG: Kyoto Encyclopedia of Genes and Genomes, MAPK: Mitogen-Activated Protein Kinase, MDA: Malondialdehyde, MF: Molecular Function, MMP: Matrix Metallopeptidase, MMP25: Matrix Metallopeptidase 25, mRNA: Messenger RNA, NF-κB: Nuclear Factor Kappa-Light-Chain-Enhancer of Activated B cells, NMSC: Non-Melanoma Skin Cancer, NOX: NADPH Oxidase, NOXO1: NADPH Oxidase Organizer 1, NR4A1: Nuclear Receptor Subfamily 4 Group A Member 1, Nrf2: Nuclear Factor Erythroid 2–Related Factor 2, ns: Not significant, PBS: Phosphate Buffered Saline, PCR: Polymerase Chain Reaction, PI: Propidium Iodide, PI3K: Phosphoinositide 3-Kinase, PIK3R1: Phosphoinositide-3-Kinase Regulatory Subunit 1, POMC: Pro-Opiomelanocortin, PPIA: Peptidylprolyl Isomerase A, p53: Tumor protein p53, R: Reverse, RAS: Rat Sarcoma virus, RIN: RNA Integrity Number, RNA: Ribonucleic Acid, RNA-seq: RNA sequencing, ROS: Reactive Oxygen Species, RT: Reverse Transcription, RTKs: Receptor Tyrosine Kinases, RT-qPCR: Quantitative Reverse Transcription Polymerase Chain Reaction, SD: Standard Deviation, STAT3: Signal Transducer and Activator of Transcription 3, TNF: Tumor Necrosis Factor, TNF-α: Tumor Necrosis Factor Alpha, TRAIL: TNF-Related Apoptosis-Inducing Ligand, TPM: Transcripts Per Million, UVA: Ultraviolet A, UVB: Ultraviolet B, α-MSH: Alpha-Melanocyte-Stimulating Hormone
Acknowledgments
None.
Author’s contributions
Shuo Xu: Formal analysis, Data curation, Data interpretation, Writing-original draft. Peixia Zhang: Supervision, Data analysis. Lijuan Qiao: Supervision. Guoping Shen and Derui Zhu: Bioinformatics technical support. Yongchun Li: Project administration, Funding acquisition. All authors read and approved the final version of the manuscript.
Funding
This research was funded by the Qinghai Science and Technology Department (2024-ZJ-937).
Availability of data and materials
Data and materials used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Declaration of generative AI and AI-assisted technologies in the writing process
The authors declare that they have not used generative AI (a type of artificial intelligence technology that can produce various types of content including text, imagery, audio and synthetic data. Examples include ChatGPT, NovelAI, Jasper AI, Rytr AI, DALL-E, etc) and AI-assisted technologies in the writing process before submission.
Competing interests
The authors declare that they have no competing interests.

